Session Information
Date: Wednesday, June 7, 2017
Session Title: Neuropharmacology
Session Time: 1:15pm-2:45pm
Location: Exhibit Hall C
Objective: IBM Watson for Drug Discovery (WDD) is a platform that analyzes vast quantities of scientific data to help researchers identify hidden connections and formulate evidence-based hypotheses. We applied WDD to discover compounds with potential for repurposing to L-DOPA-induced dyskinesia (LID).
Background: Current treatment options for LID are limited. Repurposing compounds with regulatory approval is an attractive approach to accelerate availability of novel therapeutic options. WDD uses cognitive capabilities to extract domain specific text features from published literature and identify new connections between entities of interest, including drugs. We used WDD to analyze published abstracts of a set of training compounds known to prevent LID, and applied machine learning to rank a candidate set of drugs according to similarity of linguistic context.
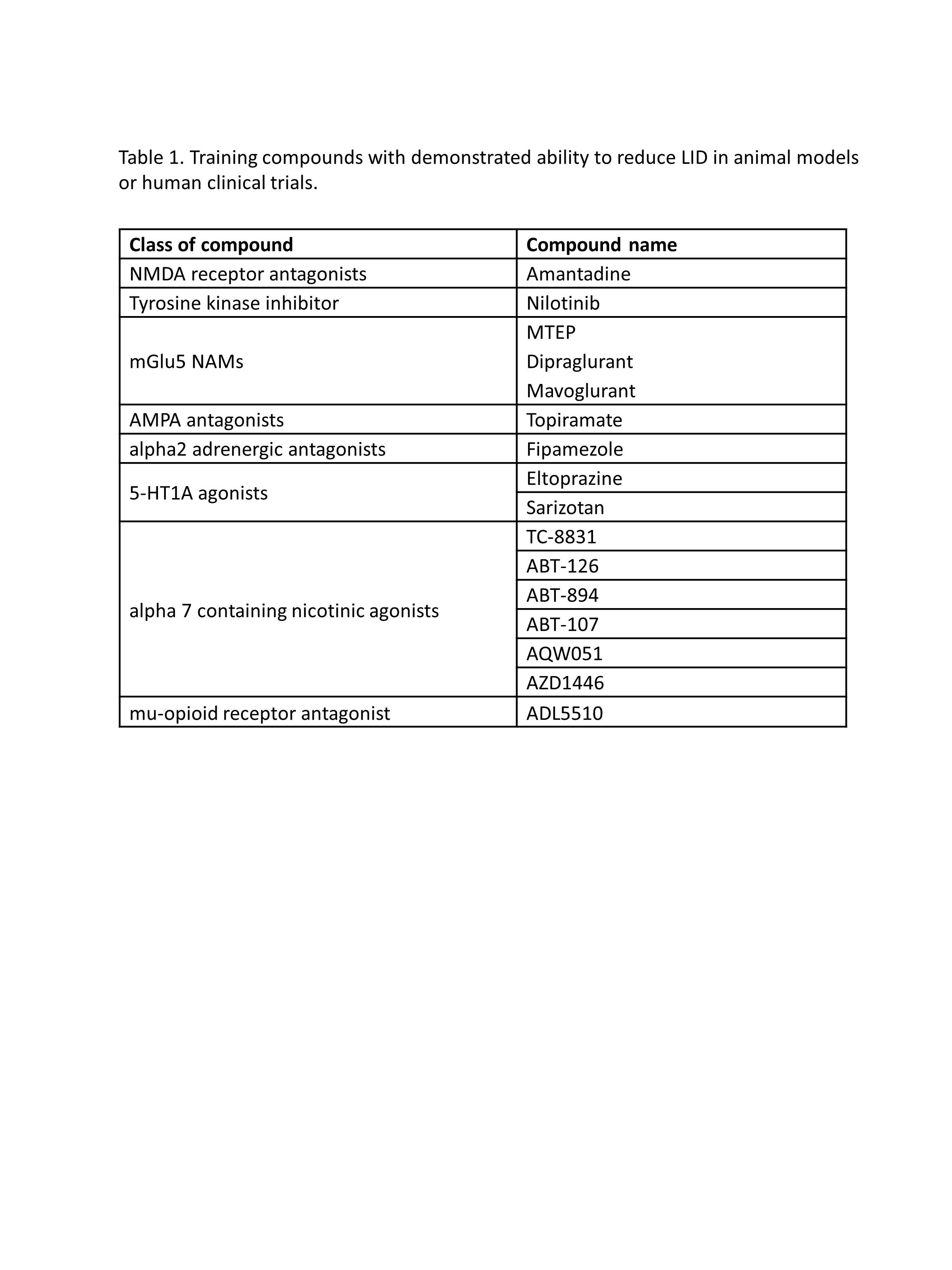
Methods: 16 training compounds were identified, via literature search, with demonstrated ability to reduce LID in animal models or human clinical trials (Table 1). Candidate drugs were filtered from 905 FDA-approved drugs with binding data available (https://www.bindingdb.org/bind/ByFDAdrugs.jsp) cross referenced with the human brain proteome (http://www.proteinatlas.org/humanproteome/brain). Dopamine agonists, antipsychotics and drugs with <5 published abstracts were also removed, leaving 385 final candidates. WDD analyzed ~450,000 Medline abstracts to generate a semantic fingerprint for each compound and then, using machine learning, create a predictive model to rank candidate compounds based on semantic similarity to the training set.
Results: Leave-one-out cross-validation demonstrated a strong predictive power of training entities over each other compared to candidates, confirming that highly ranked candidates share many properties with the training set. The top 50 ranked candidates comprised 20 compounds with preliminary evidence of antidyskinetic action with several not fully explored, 2 untested compounds with plausible antidyskinetic mechanisms of action and 6 compounds with as yet unexplored mechanisms of predicted antidyskinetic action.
Conclusions: We have employed a powerful text mining approach using WDD to identify drugs predicted to have the potential to prevent LID. Critical validation of the most promising novel compounds will be performed in rodent and non-human primate models of LID.
References: The authors would like to acknowledge: the Ontario Brain Institute and the Government of Ontario for providing access and training to the IBM Watson Drug Discovery platform.
To cite this abstract in AMA style:
N. Visanji, A. Lacoste, S. Ezell, S. Spangler, E. Argentinis, J. Brotchie, A. Lang, T. Johnston. Using cognitive computing to identify compounds with antidyskinetic potential for Parkinson’s disease [abstract]. Mov Disord. 2017; 32 (suppl 2). https://www.mdsabstracts.org/abstract/using-cognitive-computing-to-identify-compounds-with-antidyskinetic-potential-for-parkinsons-disease/. Accessed December 14, 2025.« Back to 2017 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/using-cognitive-computing-to-identify-compounds-with-antidyskinetic-potential-for-parkinsons-disease/