Session Information
Date: Monday, October 8, 2018
Session Title: Parkinson's Disease: Pathophysiology
Session Time: 1:15pm-2:45pm
Location: Hall 3FG
Objective: To investigate the gut microbiome and metabolome in Parkinson’s disease (PD).
Background: Recent evidence revealed that gut microbes promote alpha-synuclein pathology and motor deficits in a PD animal model[1]. Several case-control studies reported alterations of gut microbiome in PD patients[2], but none explored the corresponding gut metabolome, which is indicative of bacterial function and potentially a more meaningful measure of microbiome health.
Methods: We recruited 104 PD patients and 96 age-matched spouses/siblings (to control for confounding factors such as lifestyle, diet and housing condition). Demographic data and detailed medical history including medications, diet and constipation severity were collected. Stool DNA was extracted and amplicon sequencing targeting the V3-V4 region of the microbial 16SrRNA gene was performed on Illumina Hiseq platform. Stool metabolomics were analyzed using nuclear magnetic resonance spectroscopy.
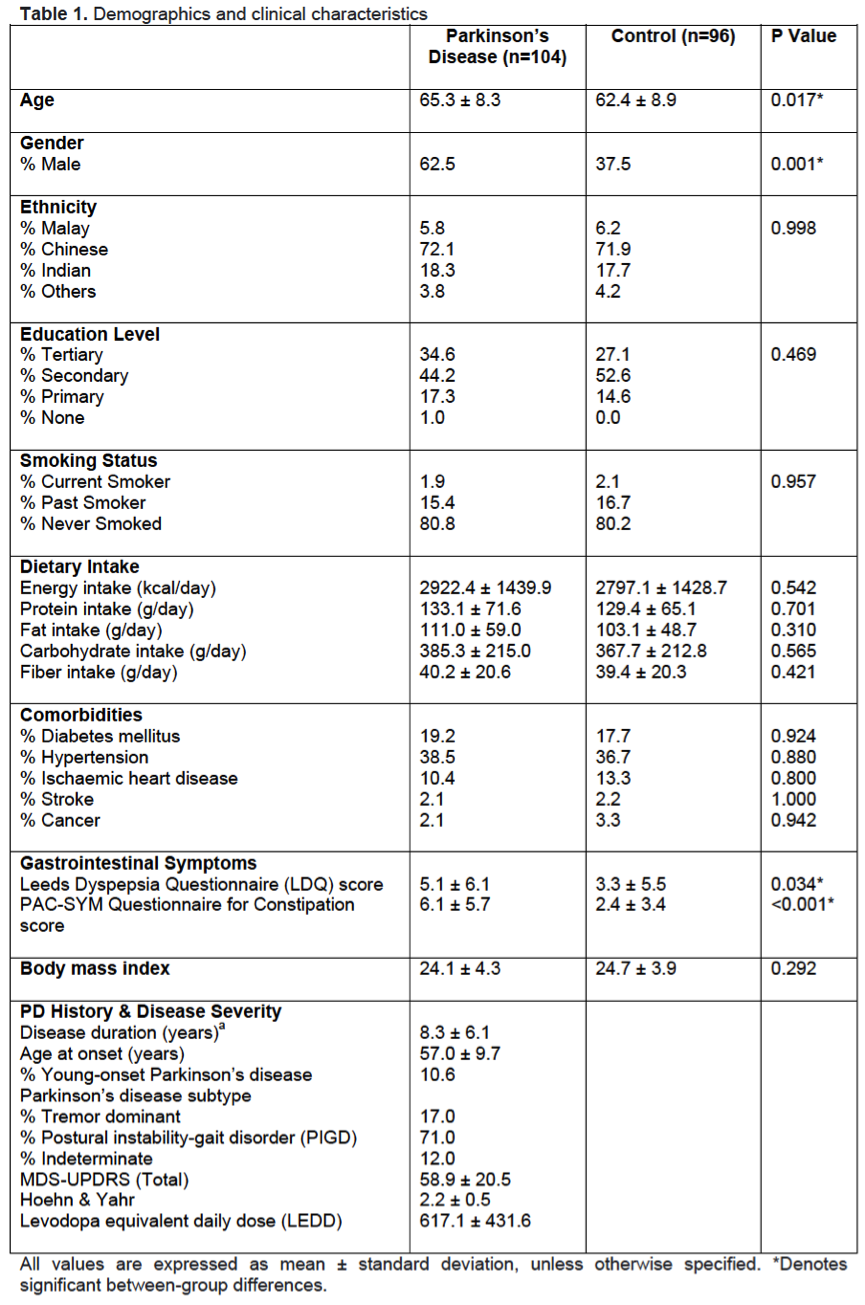
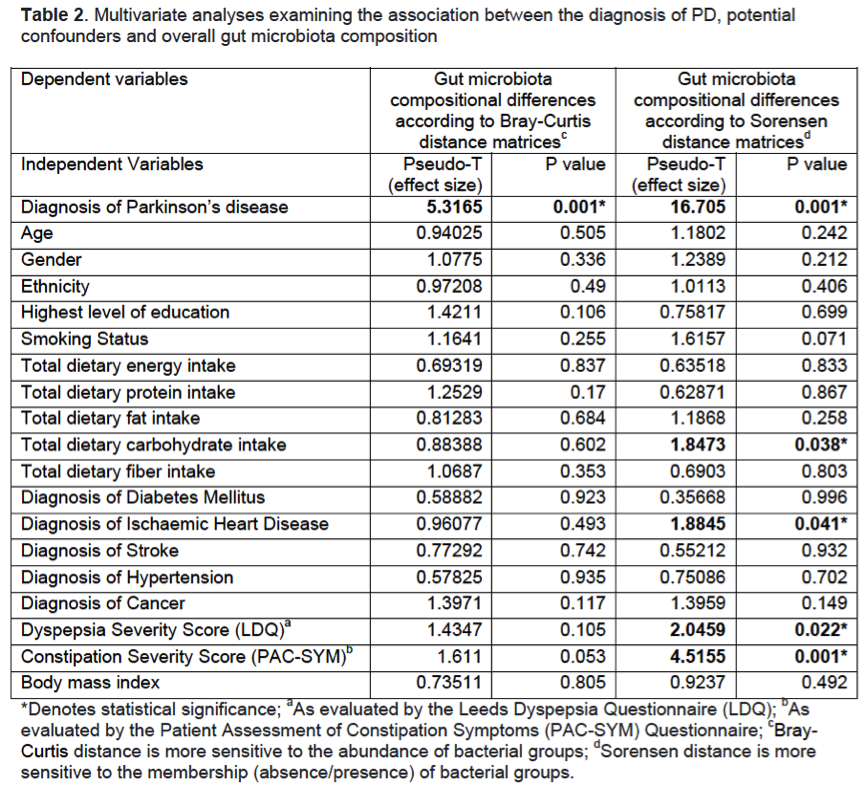
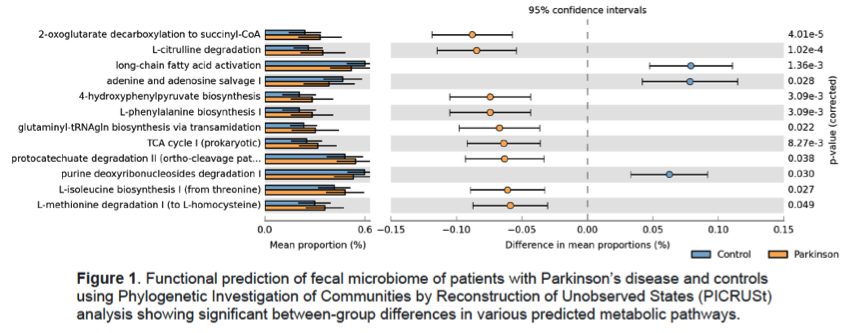
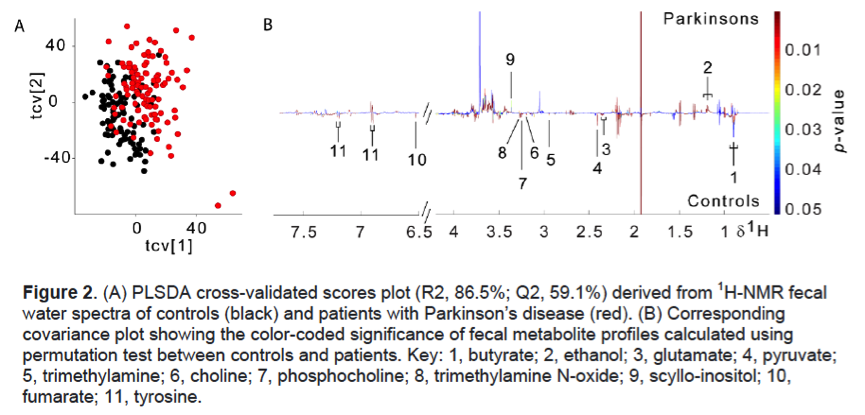
Results: PD patients were slightly older (65.3±8.3 vs 62.4±8.9, p=0.017), with a higher prevalence of males (62.5% vs 37.5%, p=0.001), and had worse dyspepsia and constipation severity scores compared to controls [Table 1]. There were no significant group differences in education level, ethnicity, smoking history, dietary pattern, comorbidities and body mass index. After controlling for potential confounders, fecal microbiota composition remained significantly different between PD and controls [Table 2] and the diagnosis of PD was significantly associated with reduced abundances of the genera Bacteroides, Parasutterella, Collinsella, Escherichia/Shigella, Prevotella and Bifidobacterium. Predicted functional metagenomics revealed alterations in energy, lipid, amino acid and purine metabolism [Figure 1]. Fecal metabolomics clearly differed between the two groups [Figure 2]. PD patients had significantly lower levels of fecal short-chain fatty acid (butyrate), amino acids (glutamate and tyrosine), derivatives of choline metabolism (choline, phosphocholine, trimethylamine, trimethylamine N-oxide) and energy metabolism (pyruvate, fumarate), but higher levels of glycolysis products (ethanol and scyllo-inositol). The specific metabolites were significantly associated with the bacterial genera on sparse partial least square model.
Conclusions: We found significant alterations in the composition and the collective function of gut microbiota in PD. Experimental models are needed to further explore the various altered gut metabolic pathways in PD.
References: 1. Sampson TR, Debelius JW, Thron T, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s Disease. Cell 2016;167:1469-1480. 2. Scheperjans F. The prodromal microbiome. Movement Disorders;2018:33:5-7.
To cite this abstract in AMA style:
A.H. Tan, C.W. Chong, C.S.J. Teh, I.K.S. Yap, M.F. Loke, J. Bowman, S.L. Song, J.Y. Tan, B.H. Ang, Y.Q. Tan, H.S. Yong, A.E. Lang, S. Mahadeva, S.Y. Lim. Unveiling the function of altered gut microbiota composition in Parkinson’s disease [abstract]. Mov Disord. 2018; 33 (suppl 2). https://www.mdsabstracts.org/abstract/unveiling-the-function-of-altered-gut-microbiota-composition-in-parkinsons-disease/. Accessed April 18, 2025.« Back to 2018 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/unveiling-the-function-of-altered-gut-microbiota-composition-in-parkinsons-disease/