Session Information
Date: Saturday, October 6, 2018
Session Title: Neuroimaging (Non-PD)
Session Time: 1:45pm-3:15pm
Location: Hall 3FG
Objective: 1. To evaluate and optimize subcortical accuracy of commonly used nonlinear deformation algorithms. 2. To optimize accuracy of automated atlas-based segmentation of the two most commonly used target regions for deep brain stimulation (DBS), the nucleus subthalamicus (STN) and the internal pallidum (GPi). 3. To guide clinicians as to which preoperative MRI sequences to acquire in patients undergoing DBS.
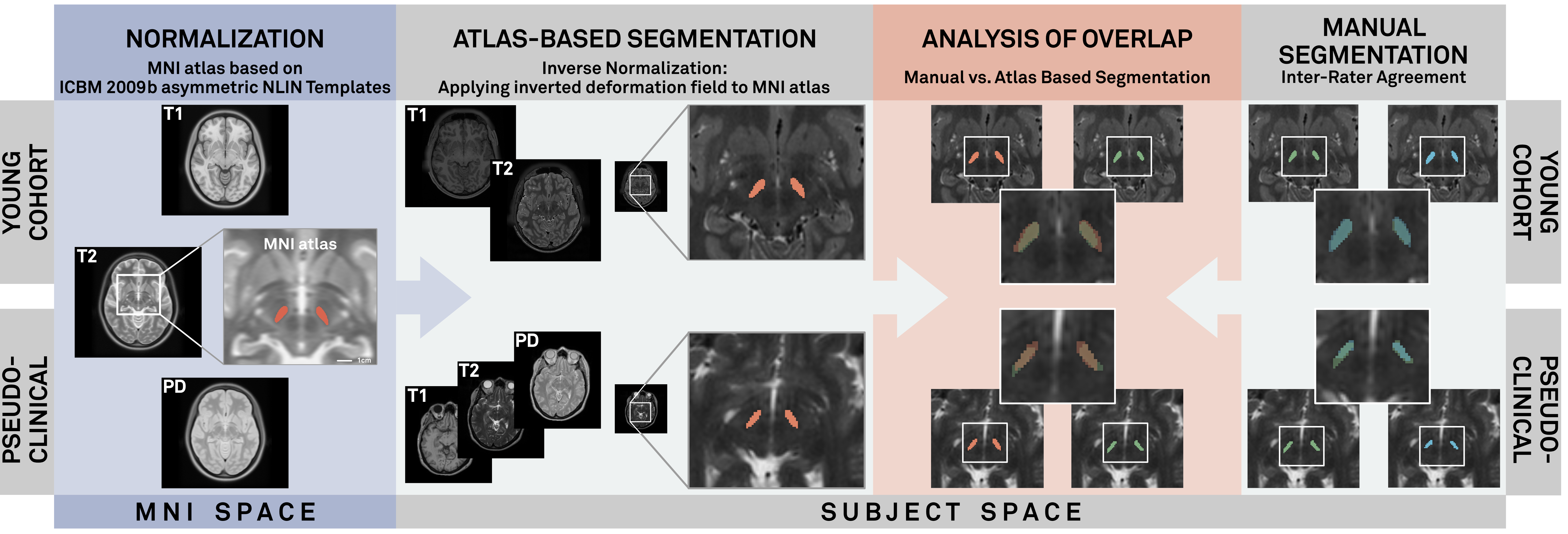
Background: Translating single-subject imaging into a common reference frame such as the MNI space is a “core concept within the field of brain mapping”. Atlases allow for delineation of DBS targets that are imperfectly resolved with clinical MRI imaging. The use of a common reference frame such as the MNI space, also allows one to leverage the vast brain imaging data available in publicly available repositories. To be useful, single-subject imaging must be accurately transformed into the atlas space. Here, we evaluated and optimized six commonly used deformation algorithms with different presets and MRI sequences and compared each outcome to manually labeled brain regions of 103 brains.
Methods: Algorithms evaluated were: New Segment, DARTEL and SHOOT (SPM); FNIRT (FSL); SyN and BSpline (ANTs). Target space was the MNI 2009b NLIN space. An atlas of the STN and GPi in MNI space was transformed to native space by inverting each deformation matrix. The resulting atlas-based segmentations were then compared to expert manual segmentations of the native brains. Overlap between the two was quantified using the Dice coefficient, mean surface distance and correlation of volumes. Two open source datasets were used: The IXI-dataset (www.brain-development.org/ixi-dataset/) and the HCP-dataset (www.humanconnectomeproject.org/).
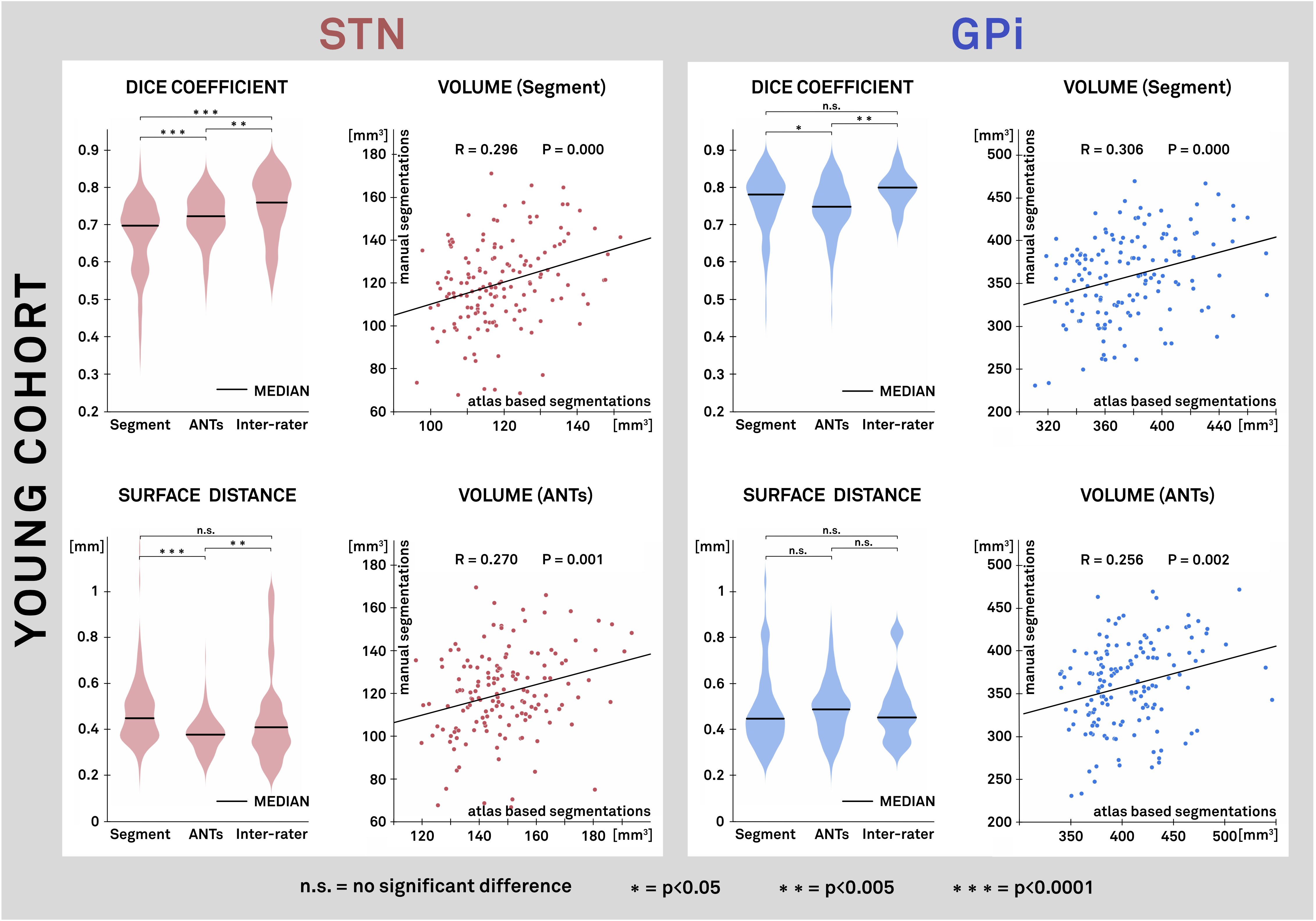
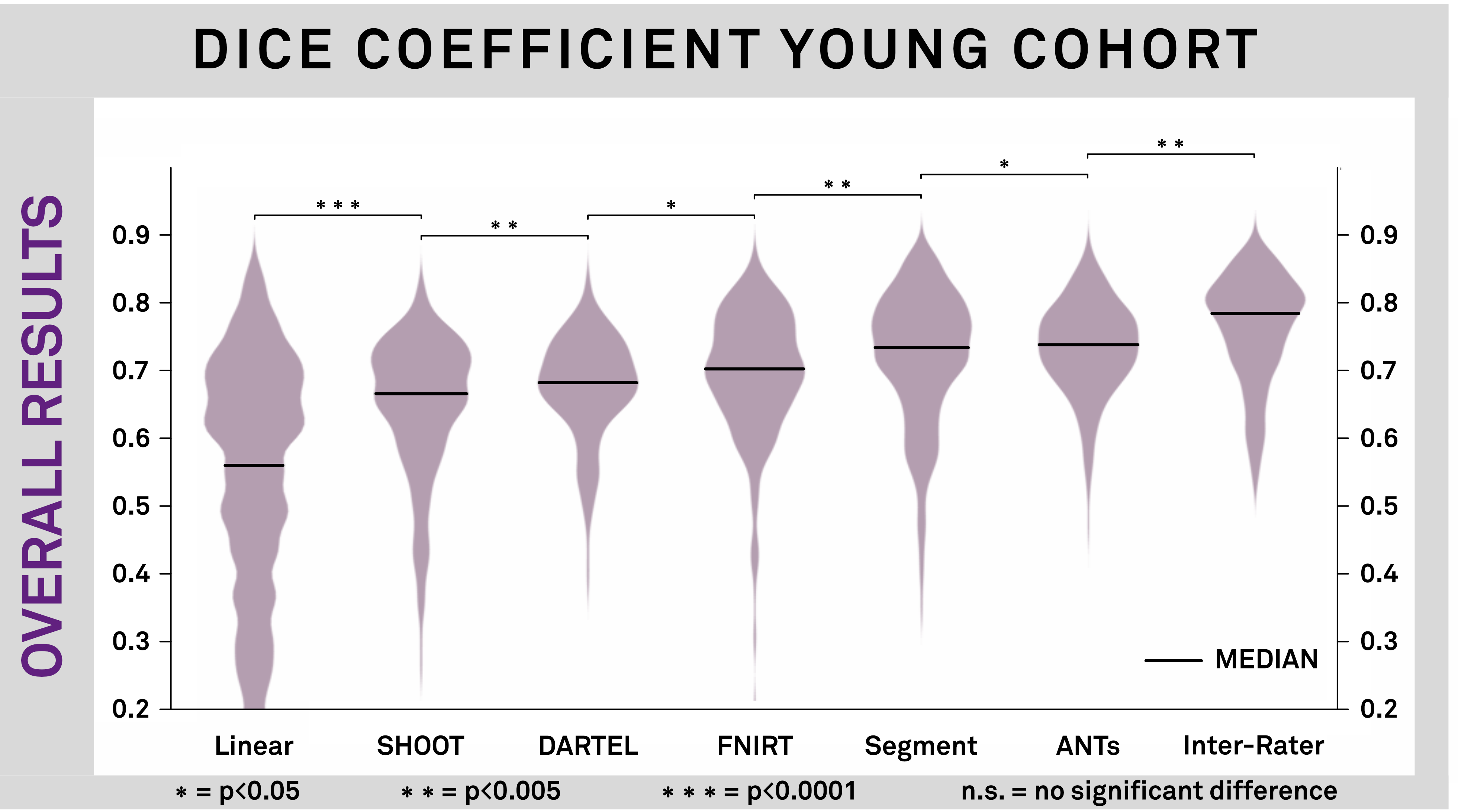
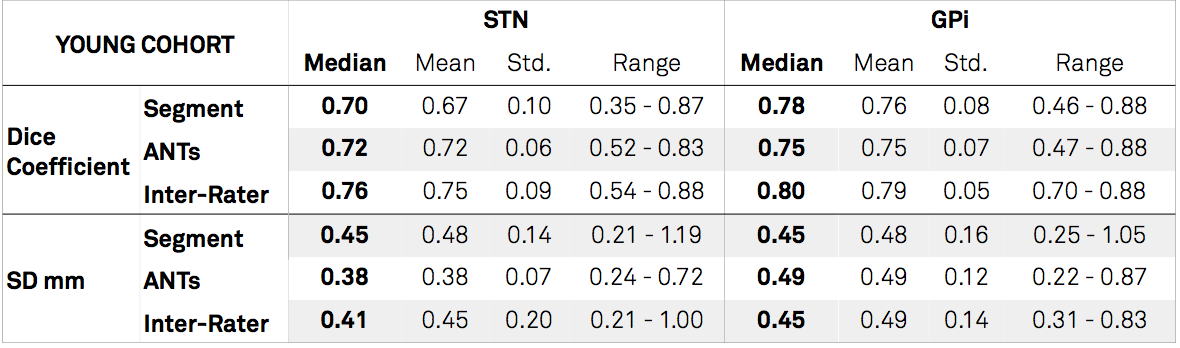
Results: Figure 2 shows the overall results for all evaluated methods by means of Dice coefficient for the Young Cohort. Performance is compared to inter-rater results of manual segmentations. The best performing algorithms were SPM New Segment with a custom tissue probability map (TPM) and ANTs SyN with an additional step to refine subcortical coregistration. Figure 3 and table 1 show the two best performing algorithms by means of Dice, mean surface distance (SD) and volume correlation for STN and GPi separately.
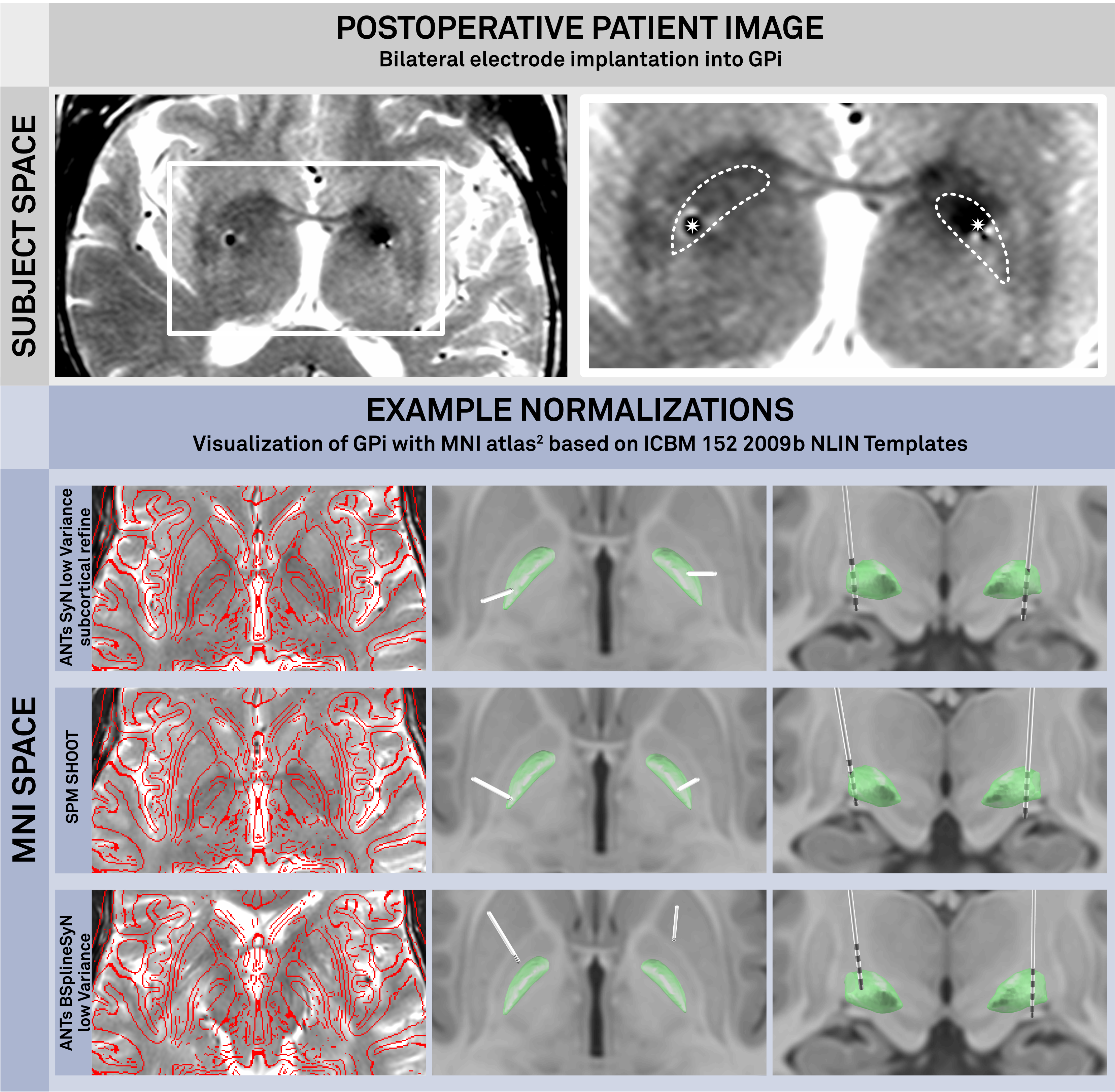
Conclusions: 1. First, we identified two nonlinear deformation algorithms that perform superiorly to other commonly used algorithms in the field. We then further optimized their performance resulting in a more precise alignment of subcortical structures. 2. The results of atlas-based segmentations are similar to inter-rater results made by expert manual raters. 3. Third, multimodal image registration was more accurate than unimodal image registration. We conclude that preoperative imaging of DBS patients should include at least a T1- and T2-weighted modality.
References: Evans, A. C. et al., 2012. Brain templates and atlases. NeuroImage. Ewert, S. et al., 2017. Toward defining deep brain stimulation targets in MNI space: A subcortical atlas based on multimodal MRI, histology and structural connectivity. NeuroImage. Horn, A. et al., 2017. Connectivity Predicts Deep Brain Stimulation Outcome in Parkinson Disease. Annals of Neurology. HCP: https://db.humanconnectome.org/ & 5. IXI: http://brain-development.org/ixi-dataset/.
To cite this abstract in AMA style:
S. Ewert, A. Horn, F. Finkel, N. Li, A. Kühn, T. Herrington. Toward optimized nonlinear deformation algorithms for subcortical DBS target regions in MRI [abstract]. Mov Disord. 2018; 33 (suppl 2). https://www.mdsabstracts.org/abstract/toward-optimized-nonlinear-deformation-algorithms-for-subcortical-dbs-target-regions-in-mri/. Accessed April 20, 2025.« Back to 2018 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/toward-optimized-nonlinear-deformation-algorithms-for-subcortical-dbs-target-regions-in-mri/