Category: Parkinson's Disease: Genetics
Objective: To identify interactions of specific pesticides with genetic loci to find causal variants for PD using gene burden analysis (BA).
Background: Pesticide exposure is consistently associated with increased risk of PD, yet most exposed individuals do not develop PD. Genetic variation individualizes susceptibility to deleterious effects of many environmental exposures, but studies of gene-environment interaction (G*E) in PD have largely relied on GWAS data that measured only common variation and had limited exposure information for suspected PD-related toxicants.
Method: We performed whole genome sequencing (WGS) on DNA from participants of the FAME study [1], a case-control study nested in the Agricultural Health Study of professional pesticide applicators and their spouses. We tested G*E using an “exposed-only” approach [2] in subgroups of male cases and controls, all of whom mixed or applied paraquat, rotenone, permethrin, or dieldrin. We used SKAT to perform BA, including weighted high impact rare and common variants to derive gene-specific burdens of variants likely to be deleterious that were enriched among cases [3]. We tested associations in two ways: 1) using all genes, and 2) using a subset of 130 a priori selected candidate genes involved in pesticide metabolism and response to oxidative stress. For significance tests of interaction we set p-value thresholds at p<1.0 x 10-5 for unselected genes and p<0.0015 for candidate genes.
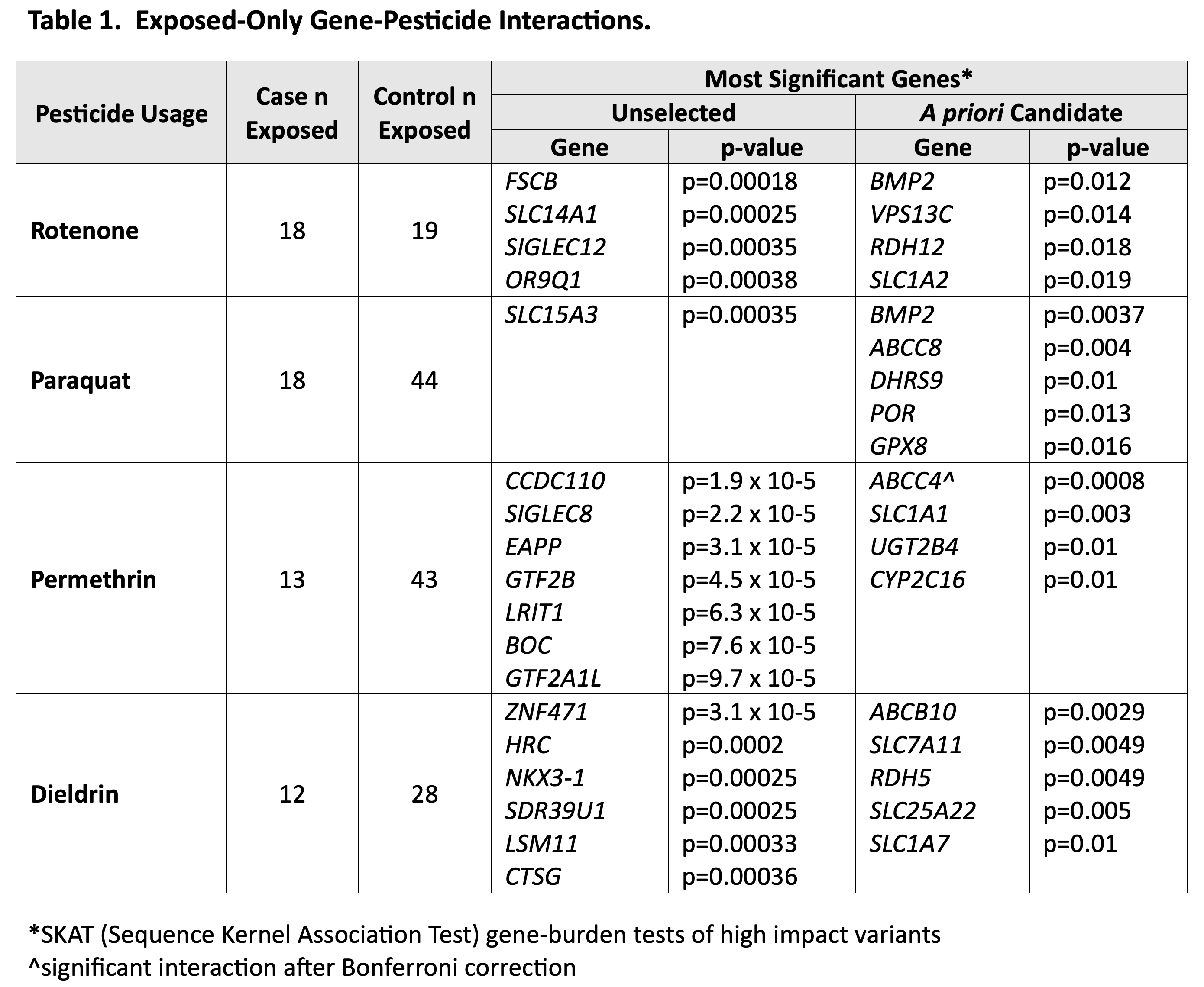
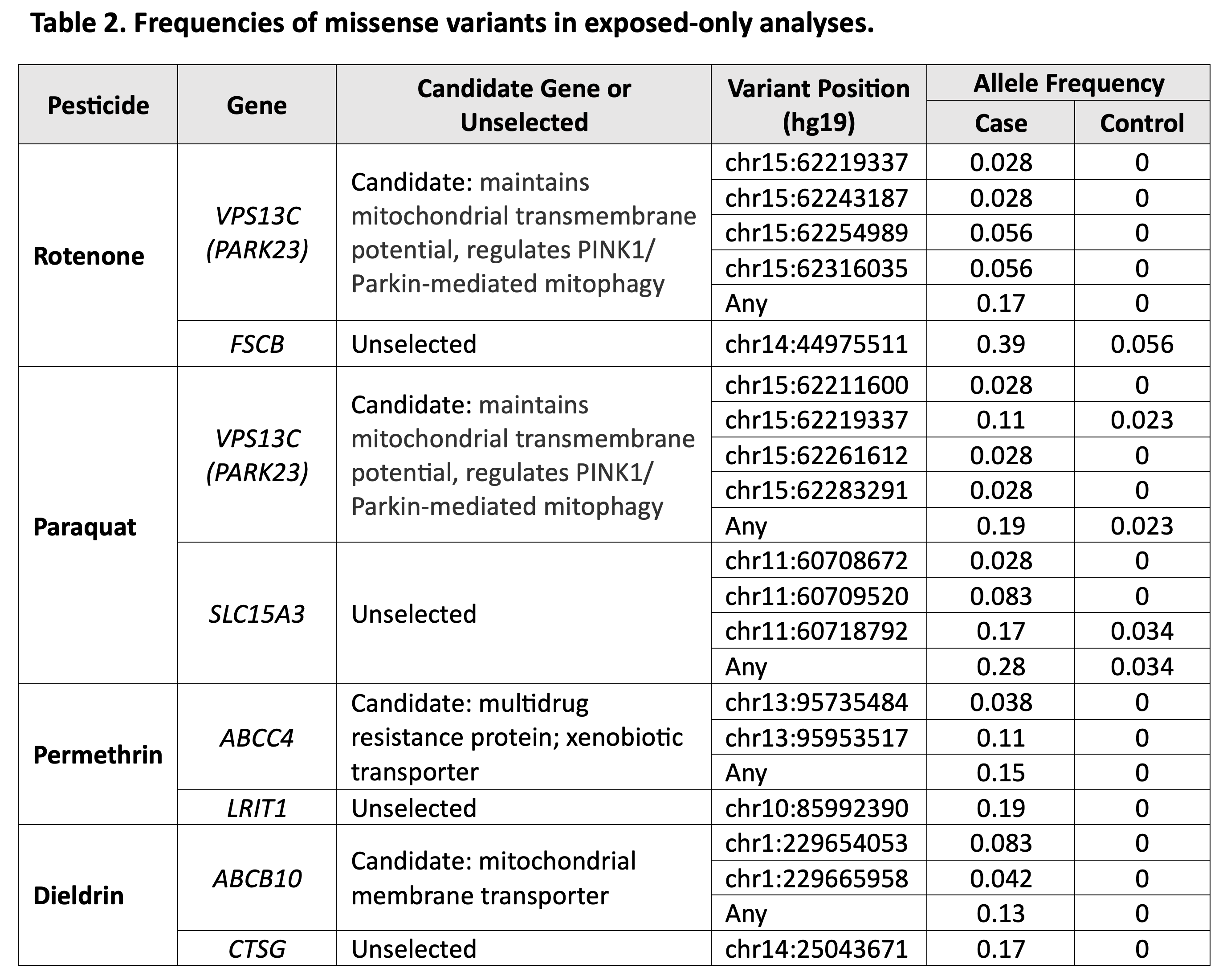
Results: We sequenced 98 cases and 239 controls overall. In exposure-specific tests of interaction (i.e., all cases and controls were exposed to that specific pesticide), ABCC4 and permethrin met the Bonferroni-corrected p-value threshold for significance. Many other interactions were near-significant despite small sample sizes.[table1] Genes with the greatest missense variant frequency disparity between cases and controls included VPS13C or FSCB with rotenone; VPS13C or SLC15A3 with paraquat; ABCC4 or LRIT1 with permethrin; and ABCB10 or CTSG with dieldrin.[table2]
Conclusion: This exposed-only analysis of whole genome high-impact genetic variation in a well-characterized cohort provides rational targets for future evaluation of genetic factors underlying individual susceptibility to pesticide-induced PD.
References: [1] Tanner CM, Kamel F, Ross GW, Hoppin JA, Goldman SM, Korell M, Marras C, Bhudhikanok GS, Kasten M, Chade AR, Comyns K, Richards MB, Meng C, Priestley B, Fernandez HH, Cambi F, Umbach DM, Blair A, Sandler DP, Langston JW. Rotenone, paraquat, and Parkinson’s disease. Environ Health Perspect. 2011 Jun;119(6):866-72.
[2] Zhao LP, Fan W, Goodman G, Radich J, Martin P. Deciphering Genome Environment Wide Interactions Using Exposed Subjects Only. Genet Epidemiol. 2015 Jul;39(5):334-46.
[3] Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin X. Sequence kernel association tests for the combined effect of rare and common variants. Am J Hum Genet. 2013 Jun 6;92(6):841-53.
To cite this abstract in AMA style:
S. Goldman, J. Kaye, M. Traglia, R. Swanson, L. Lima, S. Finkbeiner, C. Tanner. Testing gene-environment interaction in Parkinson’s disease (PD) using whole-genome sequencing of pesticide-exposed cohorts. [abstract]. Mov Disord. 2023; 38 (suppl 1). https://www.mdsabstracts.org/abstract/testing-gene-environment-interaction-in-parkinsons-disease-pd-using-whole-genome-sequencing-of-pesticide-exposed-cohorts/. Accessed December 27, 2025.« Back to 2023 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/testing-gene-environment-interaction-in-parkinsons-disease-pd-using-whole-genome-sequencing-of-pesticide-exposed-cohorts/