Category: Parkinson's Disease: Genetics
Objective: Objective
To build a multi-ancestry cohort of individuals with pathogenic variants in genes known to cause Parkinson’s disease (PD) to study monogenic PD at a global scale.
Background: The majority of monogenic PD studies were performed in European/White ancestry, severely limiting our insights into genotype-phenotype relations at a global scale. The first systematic approach embracing monogenic PD worldwide, the Micheal J Fox Foundation Global Monogenic PD (MJFF GMPD) Project, followed a systematic approach and contacted authors of publications reporting mutation carriers of variants in known PD genes [1,2]. Within the Global Parkinson’s Genetics Program (GP2) [3], the Monogenic Network (MN) took a different approach and also targeted PD centers not yet represented in the literature [4].
Method: We combined both efforts in a “merger project” to expand the cohort of monogenic PD. Centers previously included in the MJFF GMPD project were invited to participate in GP2, and previously collected samples and data were shared. Additionally, GP2’s MN expanded its aims and collected samples and data not only from individuals with unsolved but apparently monogenic PD (based on a young age at onset (AAO) or positive family history) but also with known monogenic PD. All samples underwent genotyping with the NeuroBooster array (NBA) [5].
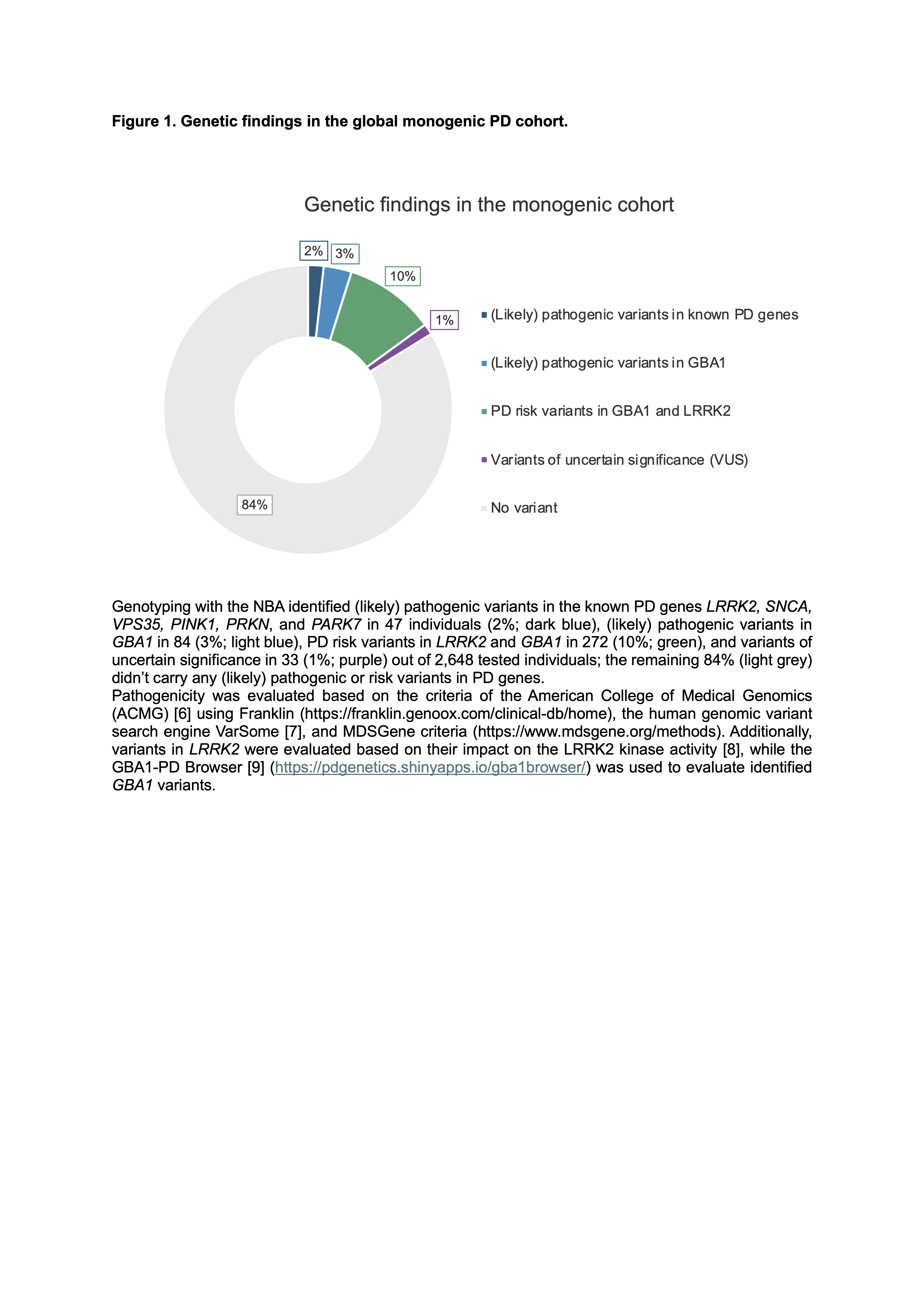
Results: Within the MN of GP2, we built a sustainable infrastructure to identify the multi-ancestry spectrum of monogenic PD. To date, 100 centers from 46 countries are included in GP2’s MN, including centers previously participating in the MJFF GMPD project. A total of 4,631 samples from affected individuals were collected. The median AAO was 49 years and 59.4% had a positive family history of PD. Genetic testing with the NBA has thus far identified pathogenic variants in 121 (including GBA1 variants), PD risk variants in LRRK2 and GBA1in 252, and variants of uncertain significance in 33 out of 2,648 tested individuals [Figure 1].
Conclusion: Integrating the MJFF GMPD project has widened the scope of GP2’s MN. Merging both projects allows for identifying the clinical and genetic spectrum of monogenic PD at a global scale and will enable investigating factors modifying monogenic PD by expanding the number of mutation carriers. Identifying patients with monogenic PD also provides the basis for recruiting individuals for future gene-specific clinical trials.
Figure 1.
References: 1. Vollstedt EJ, Kasten M, Klein C, Group MGGPsDS. Using global team science to identify genetic parkinson’s disease worldwide. Ann Neurol 2019;86(2):153-157.
2. Vollstedt EJ, Schaake S, Lohmann K, et al. Embracing Monogenic Parkinson’s Disease: The MJFF Global Genetic PD Cohort. Mov Disord 2023;38(2):286-303.
3. Global Parkinson’s Genetics P. GP2: The Global Parkinson’s Genetics Program. Mov Disord 2021;36(4):842-851.
4. Lange LM, Avenali M, Ellis M, et al. Elucidating causative gene variants in hereditary Parkinson’s disease in the Global Parkinson’s Genetics Program (GP2). NPJ Parkinsons Dis 2023;9(1):100.
5. Bandres-Ciga S, Faghri F, Majounie E, et al. NeuroBooster Array: A Genome-Wide Genotyping Platform to Study Neurological Disorders Across Diverse Populations. medRxiv 2023.
6. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17(5):405-424.
7. Kopanos C, Tsiolkas V, Kouris A, et al. VarSome: the human genomic variant search engine. Bioinformatics 2019;35(11):1978-1980.
8. Kalogeropulou AF, Purlyte E, Tonelli F, et al. Impact of 100 LRRK2 variants linked to Parkinson’s disease on kinase activity and microtubule binding. Biochem J 2022;479(17):1759-1783.
9. Parlar SC, Grenn FP, Kim JJ, Baluwendraat C, Gan-Or Z. Classification of GBA1 Variants in Parkinson’s Disease: The GBA1-PD Browser. Mov Disord 2023;38(3):489-495.
To cite this abstract in AMA style:
L. Lange, J. Junker, E-J. Vollstedt, K. Roopnarain, M. Doquenia, A. Ahmad-Annuar, M. Avenali, S. Bardien, N. Bahr, M. Ellis, C. Galandra, T. Gasser, P. Heutink, A. Illarionova, Y. Kanana, I. Keller Sarmiento, K. Kumar, S-Y. Lim, H. Madoev, I. Mata, N. Mencacci, M. Nalls, S. Padmanabhan, C. Shambetova, J. Solle, A-H. Tan, J. Trinh, E M. Valente, A. Singleton, C. Blauwendraat, K. Lohmann, Z-H. Fang, C. Klein. Studying monogenic Parkinson’s disease by building a global cohort of mutation carriers [abstract]. Mov Disord. 2024; 39 (suppl 1). https://www.mdsabstracts.org/abstract/studying-monogenic-parkinsons-disease-by-building-a-global-cohort-of-mutation-carriers/. Accessed October 22, 2025.« Back to 2024 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/studying-monogenic-parkinsons-disease-by-building-a-global-cohort-of-mutation-carriers/