Objective: To validate a recently introduced scoring algorithm predicting the diagnostic utility of exome sequencing for dystonia.
Background: Despite the increasing availability of next generation sequencing, the cost is still a major hurdle and it is important to strategies the indication of NGS in a patient of dystonia based on the clinical presentation.
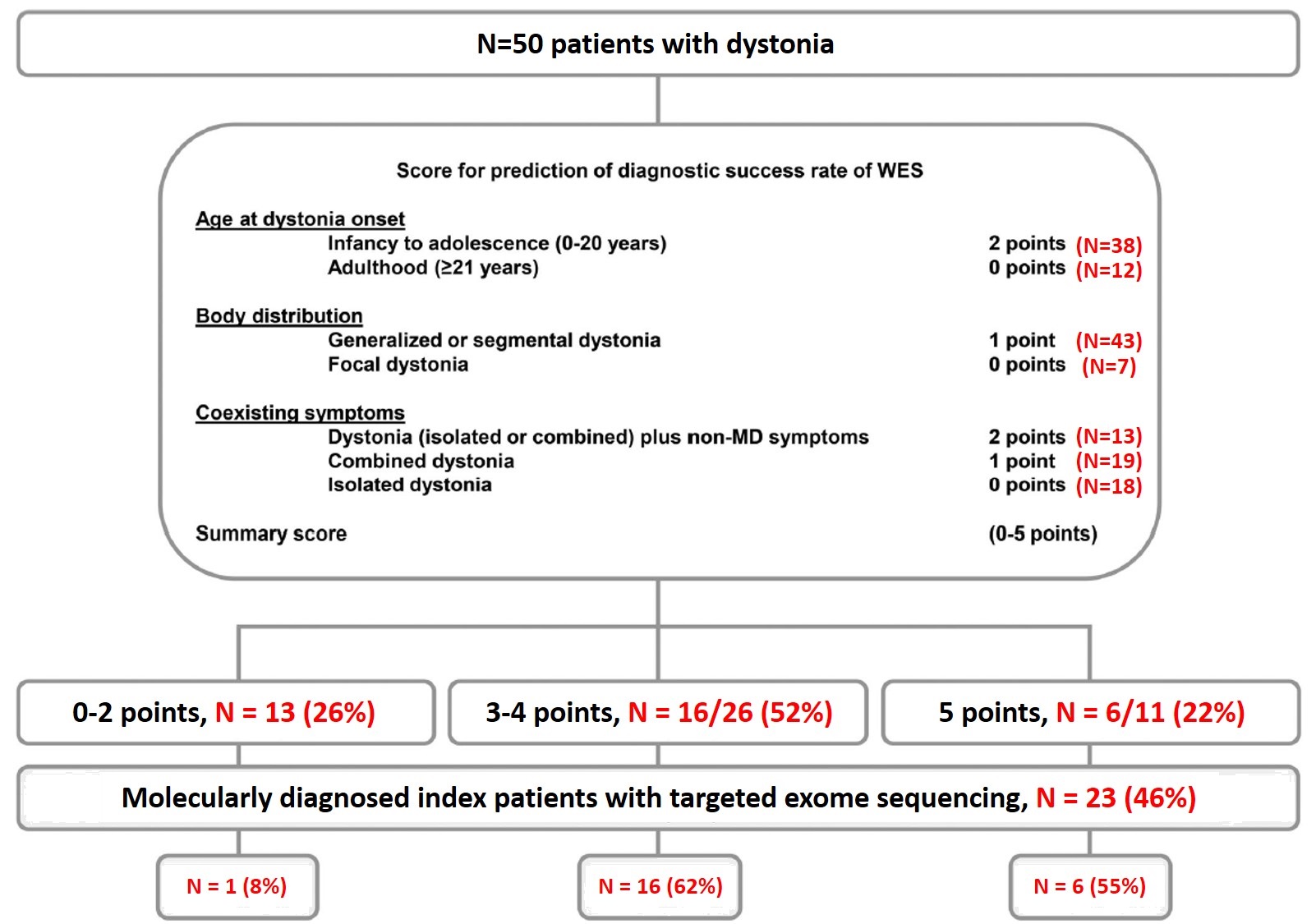
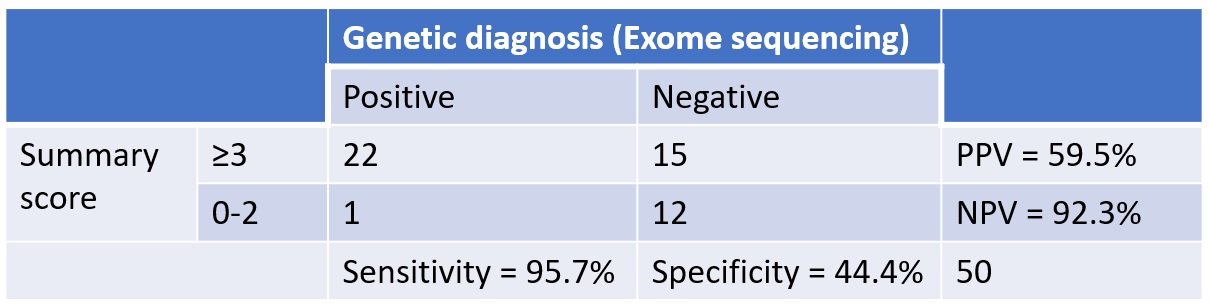
Method: By using the algorithm published by Zech et al, 2021 [1], we calculated the summary scores (0-5 points) of patients with dystonia from our database who had undergone exome sequencing. Summary scores (0-5 points) were calculated based on the age at onset (>20 years=0 points, 0-20 years=2 points), body distribution (Focal=0 point, segmental or generalized=1 point) and coexisting symptoms (Isolated=0 point, combined=1 point, isolated or combined with non-movement disorders symptoms=2 points). Sensitivity, specificity, positive and negative predictive value to predict a positive genetic diagnosis at a cut-off of 3 was calculated.
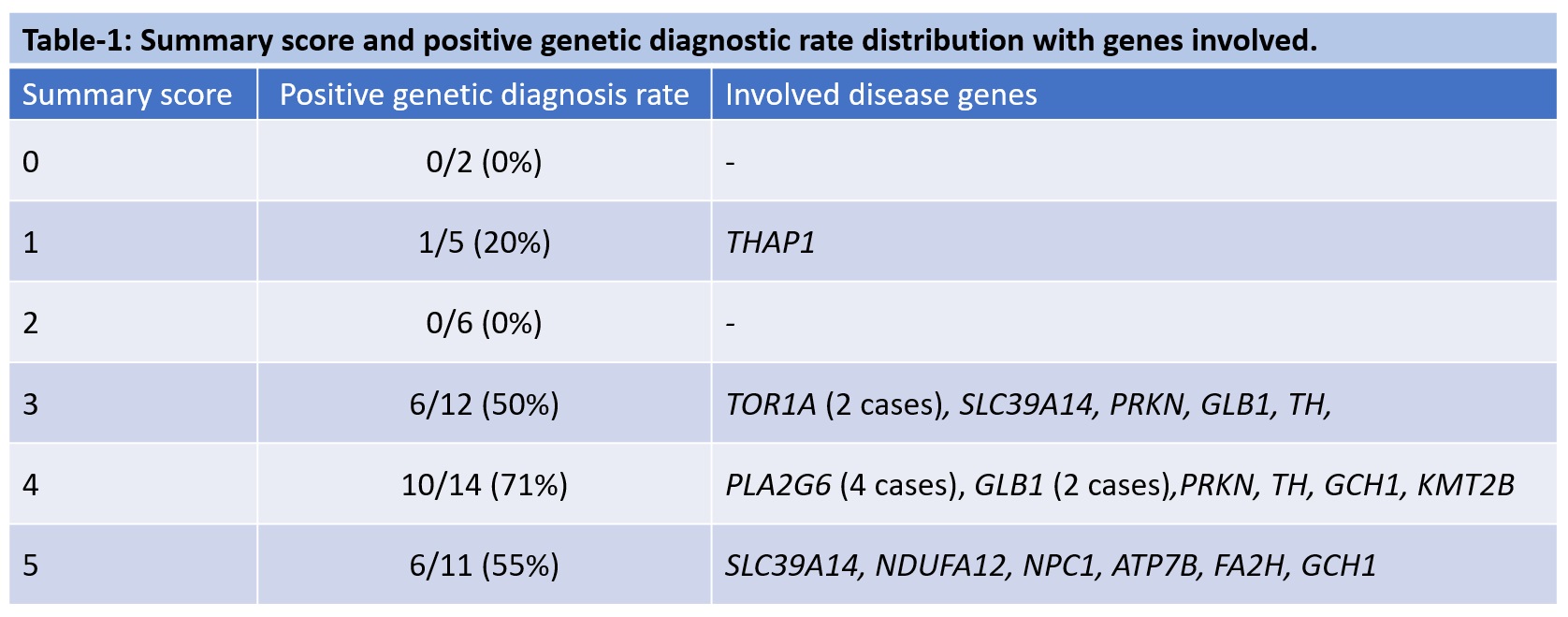
Results: 50 patients (37 males) of dystonia who had undergone exome sequencing for the evaluation of dystonia were included. The mean age at onset and presentation of the cohort was 9.1±8.81 years and 24.5±14.4 years respectively. The distribution based on the age at onset, distribution and associated symptom are shown in [Figure1]. At cut off value of 3, the sensitivity and specificity of the algorithm was 95.7% and 44.4% respectively and positive and negative predictive value was 59.5% and 92.3% respectively [Figure2]. The distribution of the summary scores and the positivity rate were as shown in [Table1]. With a score of 3 or more, close to 96% of patients with genetic positive dystonia is expected to be picked up. While a score of <3 predicts a negative testing in close to 92% patients. One patient with a summary score of 1 had a positive genetic diagnosis (DYT-THAP1). Selection bias is a major limitation in our study, as the patients were included from the database. In addition, the study population was skewed towards pediatric age group and targeted exome sequencing was used instead of whole exome sequencing.
Conclusion: The algorithm is useful in predicting the negative testing than a positive testing in our cohort. Adult onset focal dystonia such as DYT-THAP1, DYT-GNAL may be missed due to low score by the algorithm and needs to cautious in such cases.
References: [1] Zech M, Jech R, Boesch S, Škorvánek M, Necpál J, Švantnerová J et al. Scoring Algorithm‐Based Genomic Testing in Dystonia: A Prospective Validation Study. Movement Disorders. 2021 Aug;36(8):1959-64.
To cite this abstract in AMA style:
VV. Holla, K. Neeraja, A. Stezin, M. Netravathi, N. Kamble, R. Yadav, PK. Pal. Scoring Algorithm-Based Genomic Testing in Dystonia- A Validation study from a Single Centre Cohort from India [abstract]. Mov Disord. 2022; 37 (suppl 2). https://www.mdsabstracts.org/abstract/scoring-algorithm-based-genomic-testing-in-dystonia-a-validation-study-from-a-single-centre-cohort-from-india/. Accessed December 23, 2025.« Back to 2022 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/scoring-algorithm-based-genomic-testing-in-dystonia-a-validation-study-from-a-single-centre-cohort-from-india/