Session Information
Date: Sunday, October 7, 2018
Session Title: Ataxia
Session Time: 1:45pm-3:15pm
Location: Hall 3FG
Objective: To correlate differentially expressed non-coding microRNA of peripheral blood mononuclear cells (PBMCs) with SCA2 pathogenesis.
Background: Spinocerebellar ataxia aype-2 (SCA2), the most common SCA of India, is caused by expansion of uninterrupted CAG triplet repeats in first exon of ATXN2 gene. The ATXN2 is involved in the regulation of cellular processes such as RNA metabolism/stabilization, translation regulation, calcium homeostasis, and cytoskeleton reorganization etc. ATXN2 encompasses 25 exons occupying ~147 Kb of genomic space and harbouring long 3’-UTR stretch which altogether can be template for binding of several microRNAs. The non-coding miRNAs are established modifiers of gene expression in several neurological diseases but yet unexplored in SCA2.
Methods: Total RNA from PBMCs of four confirmed SCA2 patients and four matched healthy controls was analysed for differential miRNA expressions using SurePrint-Human-miRNA-Microarrays (Agilent Technologies, USA). The bioconductor “limma” package was used to calculate miRNA expression and unpaired t-test was applied to find significant (p<0.05) differential expression. Selected miRNAs were validated by qRT-PCR. Fisher’s Exact Test was used for hypergeometric distribution. Online Dianna tool was used to identify correlation of miRNAs with cellular pathways.
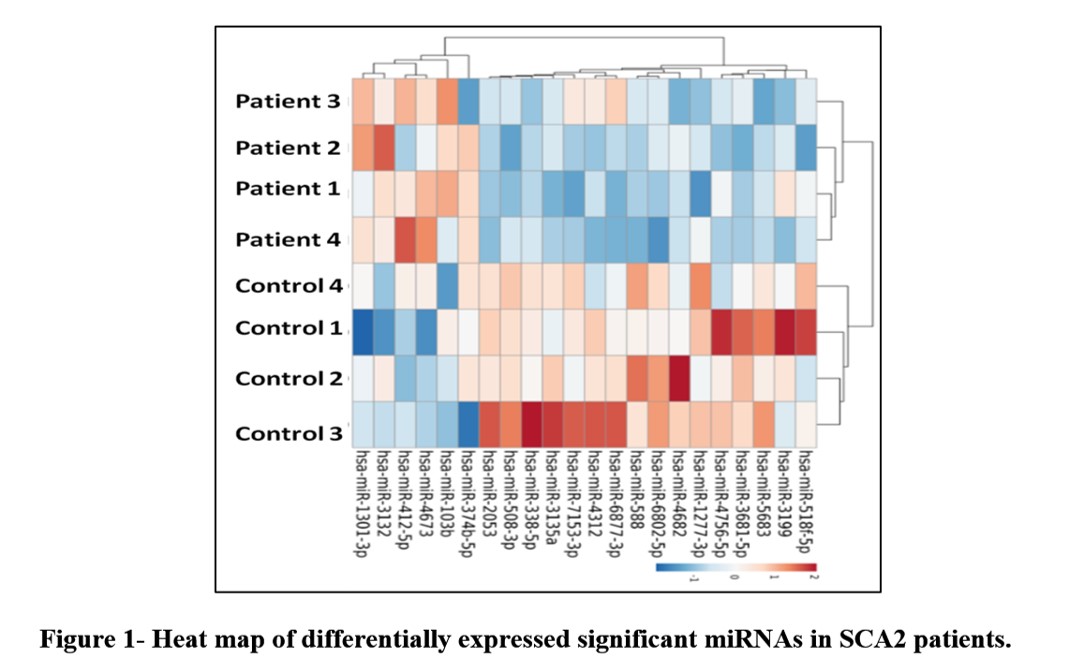
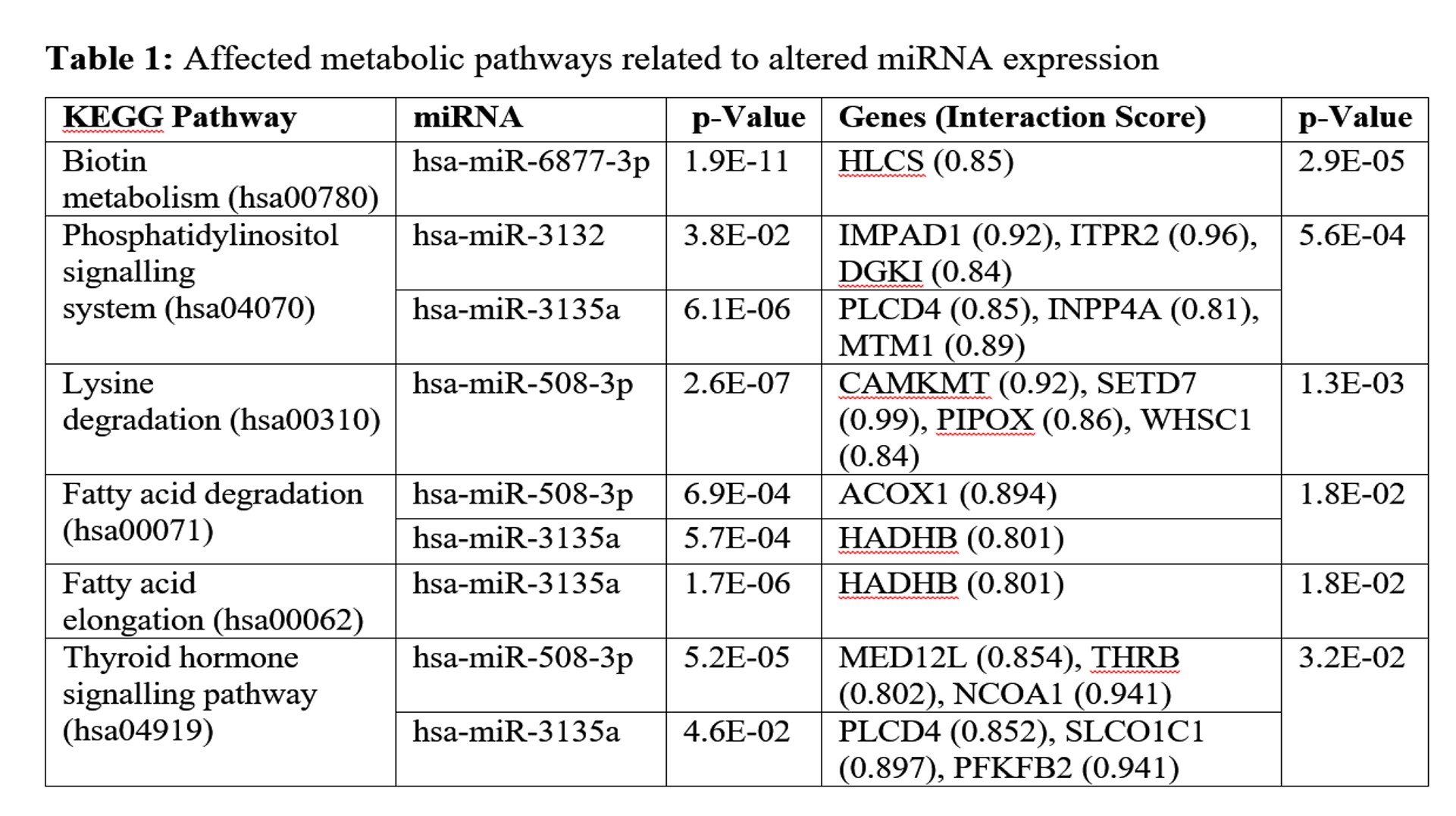
Results: Total 421 miRNA were found to be differentially expressed; 6 were upregulated and 16 were down regulated (Figure-1). Some of deregulated miRNAs were found affecting cellular pathways like, lipid metabolism, Phosphatidylinositol signalling, biotin metabolism, calcium homeostasis etc. (Table-1) Target scanning also identified genes associated with altered Ca2+ channels, IP3 pathway etc.
Conclusions: Identified miRNAs were able to demonstrate altered metabolic processes and crucial pathways associated with SCA2 pathogenesis in concordance with our earlier proteomic work and other studies. Few miRNAs can be potential biomarkers or targets to develop new therapeutic approaches.
To cite this abstract in AMA style:
V. Swarup, R. Singh, H. Singh, I. Singh, M. Faruq, S. Vivekanandhan, A. Srivastava. MicroRNAs Unveil Metabolic Imbalance in Spinocerebellar Ataxia Type-2 [abstract]. Mov Disord. 2018; 33 (suppl 2). https://www.mdsabstracts.org/abstract/micrornas-unveil-metabolic-imbalance-in-spinocerebellar-ataxia-type-2/. Accessed April 20, 2025.« Back to 2018 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/micrornas-unveil-metabolic-imbalance-in-spinocerebellar-ataxia-type-2/