Category: Parkinson's Disease: Genetics
Objective: Perform Chromosome X-Wide Association Study (XWAS) per country from The Latin American Research Consortium on the Genetics of PD (LARGE-PD) and meta-analyze the results in order to identify genetic variants on the chromosome X (chrX) associated with Parkinson’s disease (PD) risk in Latinos.
Background: It is known that males are 1.5x more likely to have PD than females. However, little is known about the impact of genetic variants on the chrX. Although classically, at least in Europeans, these studies are performed including all participants, this may not be the best approach when studying admixed individuals like Latinos.
Method: We used data from LARGE-PD composed of individuals from Brazil, Chile, Colombia, Peru and Uruguay [1]. We separated the data per country and each dataset was cleaned separately following a modified version of LeGuen et al. [2] protocol. We chose not to follow the ancestry-based removal steps of the original protocol as these steps are not feasible for admixed populations. We imputed each dataset using the TOPMed Imputation Server [3]. To avoid population structure, we run the regression by country (using PLINK 2[4]) and meta-analyzed the results (using Metal [5]). We defined the statistical significance cutoff based on the number of effective tests for the chrX [6].
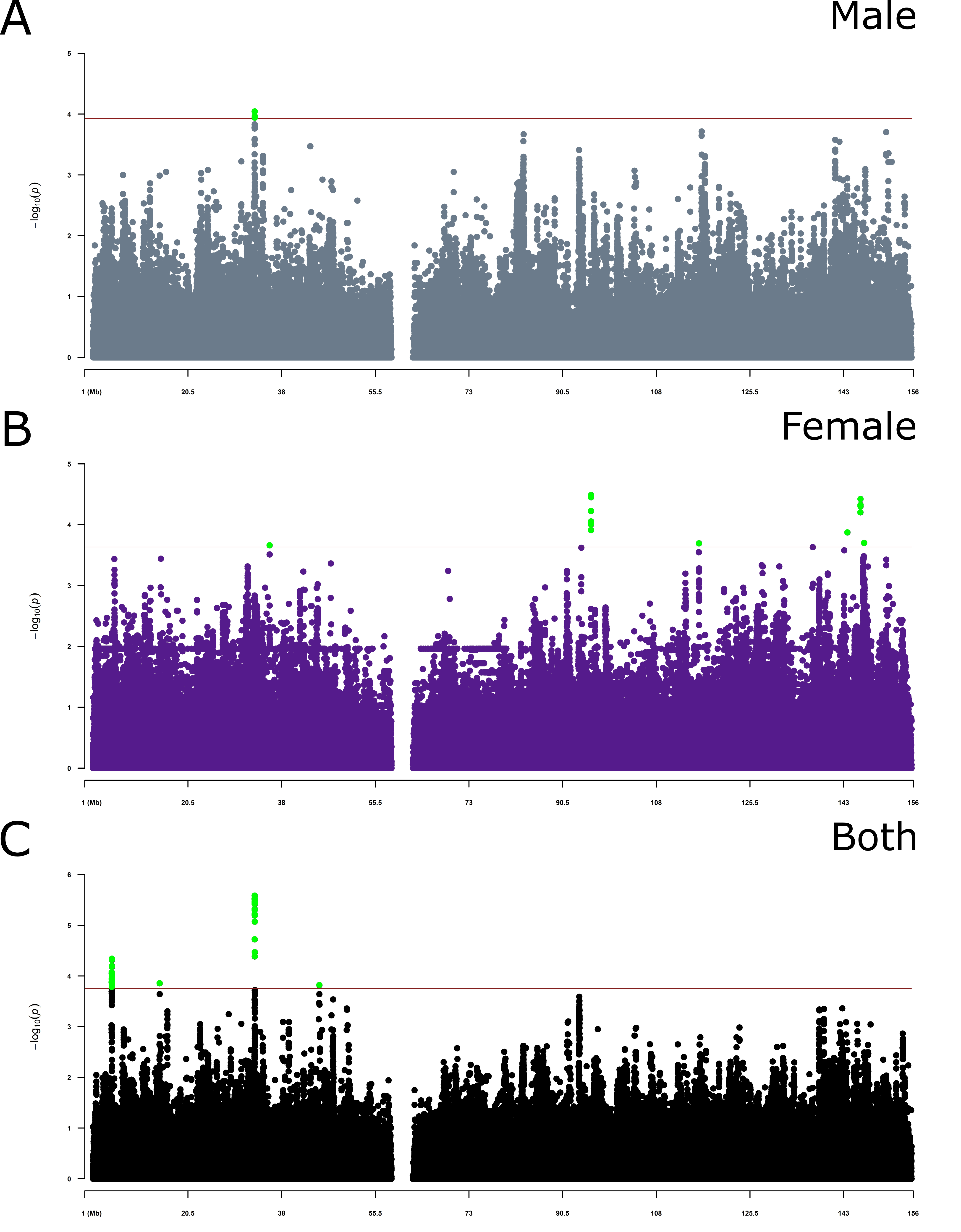
Results: We identified one region in males, four in females and four when combining both sexes significantly associated to PD risk ([figure 1]). The peak in males is inside DMD gene. One of the peaks in females is near MAGEB16 while the other peaks are in intronic regions. The four peaks when looking at both sexes are near ASB9 and NLGN4X, and inside DMD and SYN1.
Conclusion: We performed XWAS followed by meta-analysis to identify variants associated to PD risk in Latinos. We found statistical significance in DMD and SYN1 gene and near from other three genes. The DMD gene is the largest known human gene and provides instructions to produce a protein called dystrophin, a protein found primarily in skeletal and cardiac muscles, but small amounts are present in nerve cells in the brain [7]. The protein SYN1 protein is mainly associated with synaptogenesis and regulation of neurotransmitter release from presynaptic neuron terminals. Interestingly, induced neurons from PD patients carrying a SNCA mutation showed disrupted synaptic signaling and transcriptional alterations in several synaptic genes including SYN1[8].
References: [1] D.P. Loesch, A.R.V.R. Horimoto, K. Heilbron, E.I. Sarihan, et al. Characterizing the Genetic Architecture of Parkinson’s Disease in Latinos. Ann. Neurol. (2021), 10.1002/ana.26153
[2] Le Guen Y, Napolioni V, et al. Common X-chromosome variants are associated with parkinson’s disease risk. Ann Neurol 2021. https://doi.org/10.1002/ana.26051.
[3] Das, S., Forer, L., Schönherr, S, et al. Next-generation genotype imputation service and methods. Nature Genetics (2016), 48(10), 1284–1287.
[4] Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience, 4.
[5] Willer, Cristen J et al. “METAL: fast and efficient meta-analysis of genomewide association scans.” Bioinformatics (Oxford, England) vol. 26,17 (2010): 2190-1. doi:10.1093/bioinformatics/btq340
[6] Bretherton, C. S., Widmann, M., Dymnikov, V. P., Wallace, J. M., & Bladé, I.. The effective number of spatial degrees of freedom of a time-varying field. Journal of Climate (1999), 12(7), 1990–2009.
[7] Anderson, J L et al. “Brain function in Duchenne muscular dystrophy.” Brain : a journal of neurology vol. 125,Pt 1 (2002): 4-13. doi:10.1093/brain/awf012
[8] Kouroupi, G. et al. Defective synaptic connectivity and axonal neuropathology in a human iPSC-based model of familial Parkinson’s disease. Proc. Natl. Acad. Sci. USA 114, E3679–E3688 (2017).
To cite this abstract in AMA style:
T. Peixoto Leal, S. Rao, M. Inca-Martinez, M. Gouveia, J. French-Kwawu, V. Borda, E. Mason, D. Loesch, A. Horimoto, E. Sarihan, M. Cornejo-Olivas, L. Torres, P. Mazzetti, C. Cosentino, E. Sarapura-Castro, A. Rivera-Valdivia, A. Medina, E. Dieguez, V. Raggio, A. Lescano, V. Tumas, V. Borges, H. Ferraz, C. Rieder, A. Schumacher-Schuh, B. Santos-Lobato, C. Velez-Pardo, M. Jimenez-Del-Rio, F. Lopera, S. Moreno, P. Chana-Cuevas, W. Fernandez, G. Arboleda, H. Arboleda, C. Arboleda-Bustos, D. Yearout, C. Zabetian, T. Thornton, T. O'Connor, I. Mata. Meta-analysis between LARGE-PD cohorts reveals new potential susceptibility regions on the chromosome X [abstract]. Mov Disord. 2022; 37 (suppl 2). https://www.mdsabstracts.org/abstract/meta-analysis-between-large-pd-cohorts-reveals-new-potential-susceptibility-regions-on-the-chromosome-x/. Accessed December 22, 2025.« Back to 2022 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/meta-analysis-between-large-pd-cohorts-reveals-new-potential-susceptibility-regions-on-the-chromosome-x/