Objective: To address the gap existing during clinical diagnosis in differentiating among various forms of Parkinson syndrome (PS) by using mass spectrometry (MS)-based brain proteomics.
Background: PS is clinically characterized by a combination of 2 of the 3 of bradykinesia, rigidity, and tremor. Several pathological entities manifest as PS including Parkinson’s disease (PD), progressive supranuclear palsy (PSP), and multiple system atrophy (MSA). The course, response to treatment, and life expectancy are different in these disorders. Clinical distinction between these conditions is not always possible, especially during early stage and there are no biological markers that help identify a specific disorder.
Method: Patients were evaluated at Movement Disorders Clinic Saskatchewan and followed longitudinally. Autopsy is offered to all subjects at no cost. At autopsy, the brain was divided at midline, one-half frozen at -80C and the other half examined by a neuropathologist for pathological confirmation. Frozen brain tissue from patients with final diagnosis of PD, PSP, and MSA were analyzed using MS-based proteomics blinded to the diagnosis.
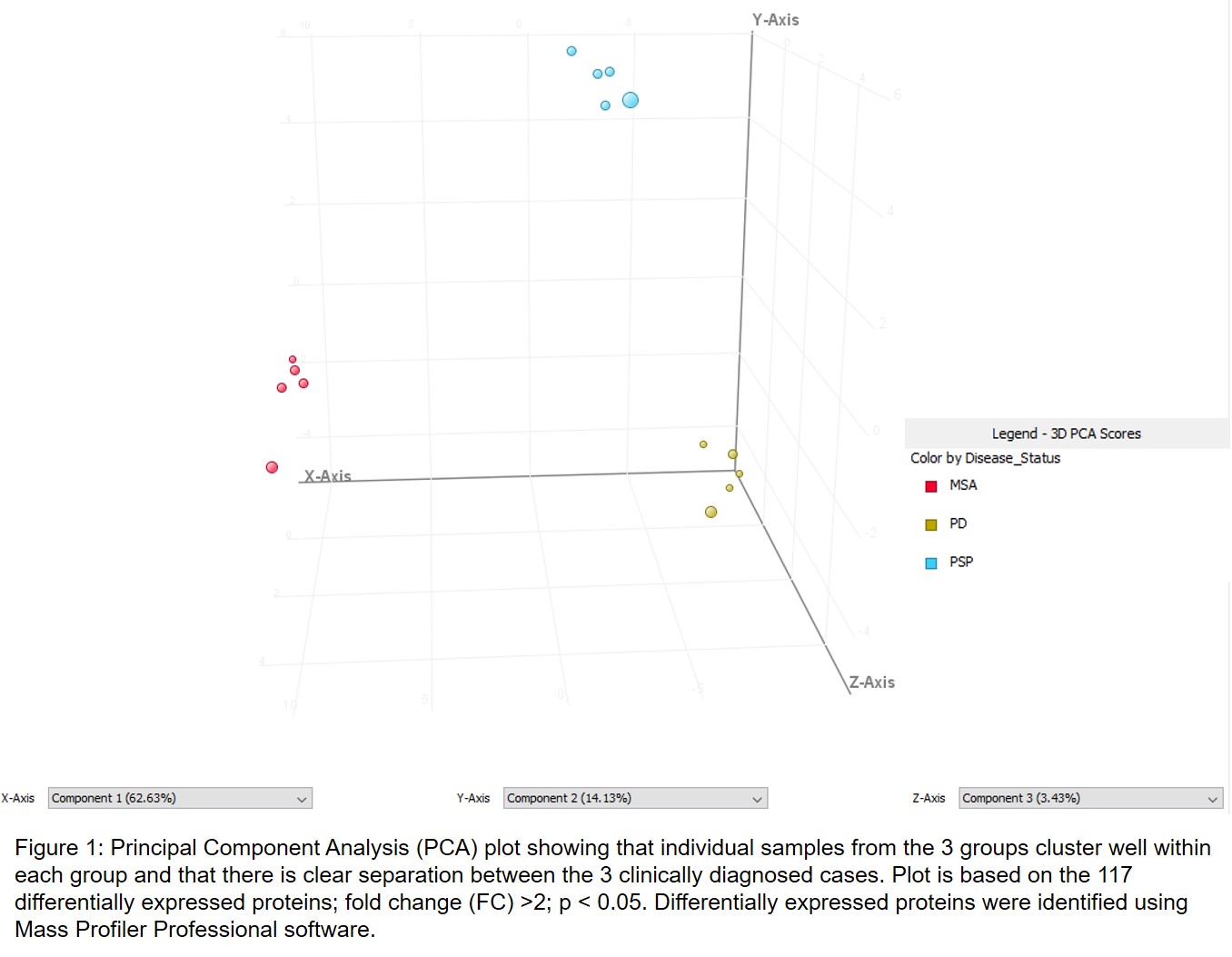
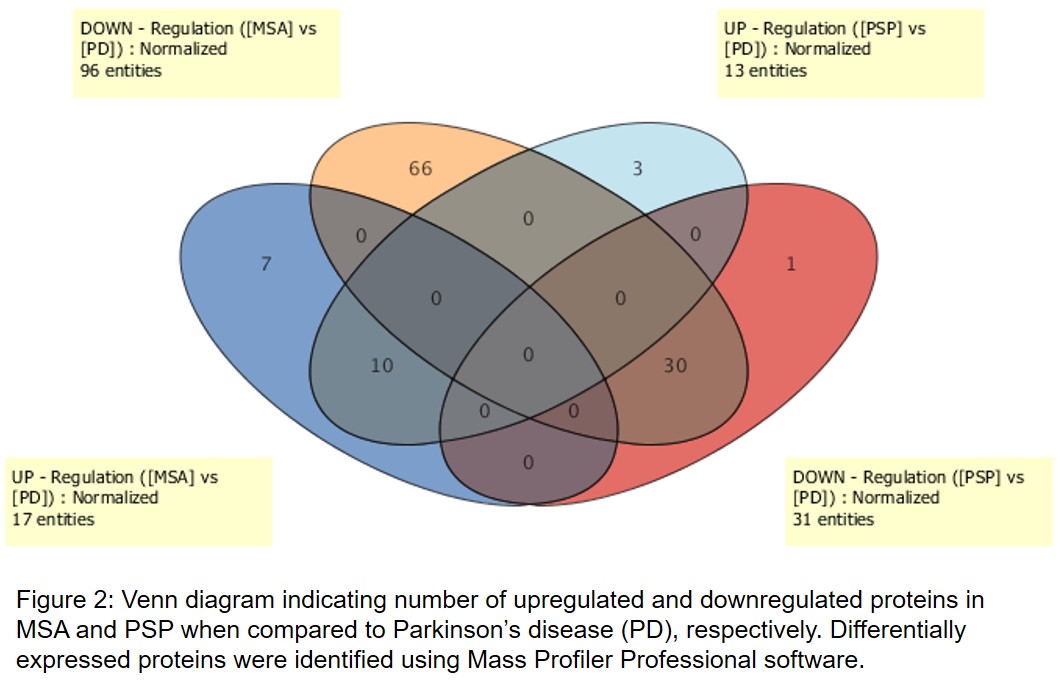
Results: Five PD, 5 PSP, and 5 MSA brains were analyzed by MS. Database search identified 7098 proteins in all samples. Statistical analysis (FC>2; p<0.05) produced 117 differentially expressed proteins. Principal component analysis showed that individual samples from the 3 groups cluster well within each group and that there is clear separation between the 3 clinically diagnosed cases. Of these proteins, 17 were found to be upregulated and 96 downregulated in MSA when compared to PD. Also, 13 proteins were found to be upregulated and 31 downregulated in PSP when compared to PD. Furthermore, 7 proteins upregulated were unique to MSA and 10 upregulated were common between MSA and PSP. Similarly, 66 proteins downregulated were unique to MSA and 30 downregulated were common between MSA and PSP. Finally, 3 proteins upregulated and 1 downregulated were unique to PSP. Marker proteins include septins, synapsin-2, brain acid soluble protein 1, MARCKS, synaptophysin, complexin-2, neuroplastin and others.
Conclusion: Our data show that MS-based protein analysis of brain can distinguish between PD, PSP, and MSA cases. This observation has a potential for future discovery of biological markers in patients with different PS variants.
To cite this abstract in AMA style:
A. Rajput, P. Chumala, B. Thompson, A. Rajput, G. Katselis. Mass spectrometry-based proteomics brain analysis to distinguish between Parkinson syndromes [abstract]. Mov Disord. 2020; 35 (suppl 1). https://www.mdsabstracts.org/abstract/mass-spectrometry-based-proteomics-brain-analysis-to-distinguish-between-parkinson-syndromes/. Accessed April 29, 2025.« Back to MDS Virtual Congress 2020
MDS Abstracts - https://www.mdsabstracts.org/abstract/mass-spectrometry-based-proteomics-brain-analysis-to-distinguish-between-parkinson-syndromes/