Category: Parkinson's Disease: Genetics
Objective: To investigate the frequency of copy number variations (CNVs) in SNCA and PRKN in a cohort of Parkinson’s disease (PD) patients and healthy controls from Kazakhstan.
Background: Little is known about the genetics of PD in Kazakhstan. In 2023, we initiated a Michael J. Fox Foundation (MJFF)-funded project titled “Developing a Collaborative Parkinson’s Disease Biobank in Central Asia and Transcaucasia (CAT-PD)”. Presently, we have DNA samples from 1,258 PD patients and 518 healthy controls from Kazakhstan, recruited and genotyped using the Illumina Neurobooster array by the Global Parkinson’s Disease Genetics Program (GP2).
Method: Raw IDAT files from genotyping were processed by the GenomeStudio v2.0.5 software. B-allele frequency and log R ratio intensity files were generated and manually inspected in 1,258 PD cases and 518 healthy controls for CNVs in SNCA and PRKN.
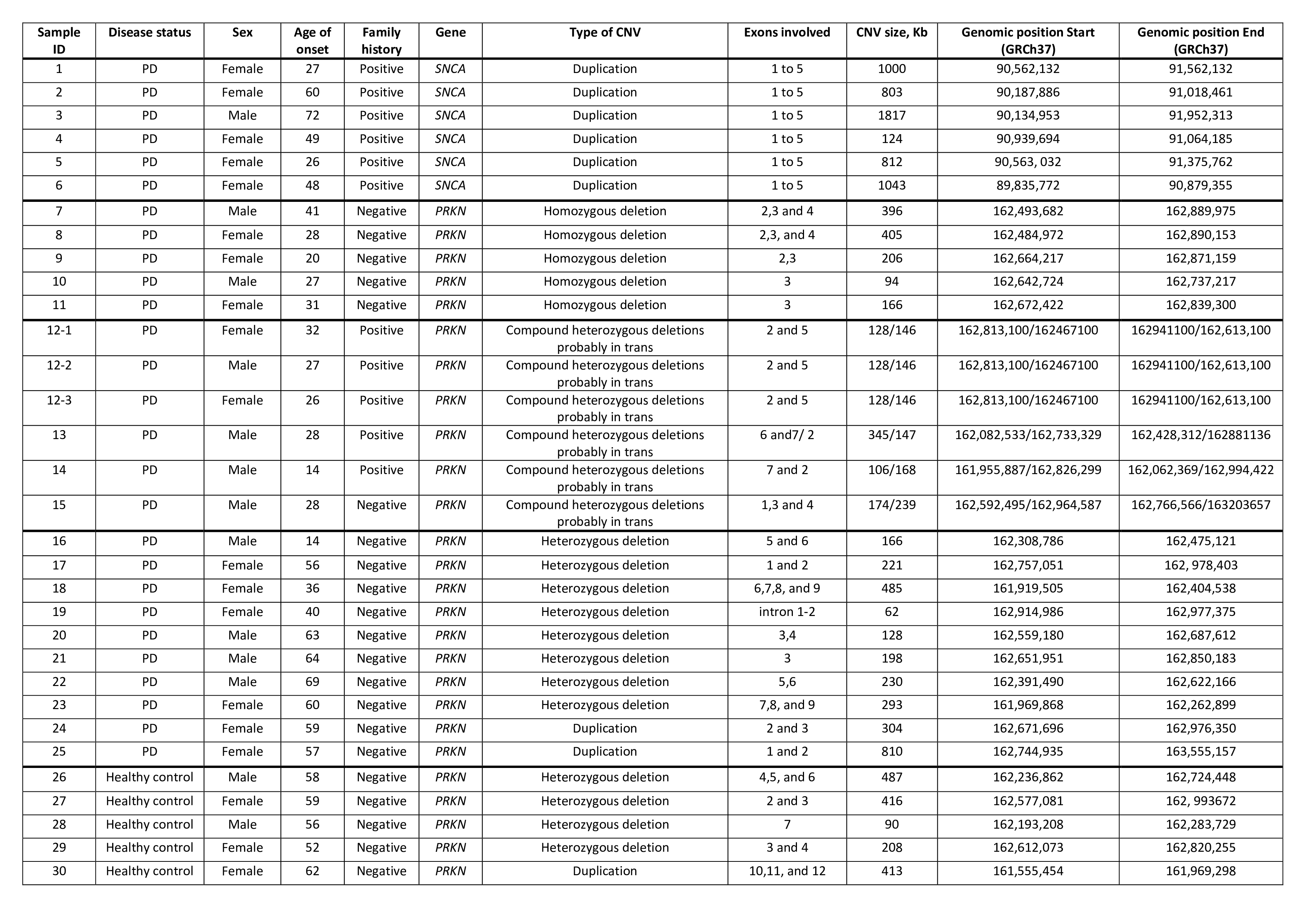
Results: The manual inspection detected CNVs in SNCA and PRKN in 32 individuals (13 males, 19 females, 3 of whom are related subjects), including 27 PD patients (2.1%) and 5 controls (0.96%), Chi-Sq, p<0.05, GRCh37 (Table 1). SNCA duplications were found in 6 PD cases with a mean onset age of 47±16 (range 26–72 years). Homozygous PRKN deletions occurred in 5 unrelated cases (mean age 29.4±6.8, range 20–41). Compound heterozygous PRKN deletions (probably in trans) were identified in 6 individuals (mean age 28.3±5.6, range 14-32, 3/6 patients from the same family with 5 affected). PRKN duplications were observed in 2 patients and 1 control (mean age 59.3±2.0). Heterozygous PRKN deletions were present in 8 patients (mean age 50±17) and 4 controls (mean age 56±2.6), Chi-Sq, p>0.05. A family history of PD was noted in 9/30 (30%) unrelated cases. PRKN deletions were spread across exons 1–9. Overall, 1,3% of the PD cohort had likely disease-causing CNVs. Patients with a single PRKN deletion did not exhibit pathogenic single nucleotide variants based on genotyping data.
Conclusion: The prevalence of CNVs in SNCA and PRKN among Kazakhstani PD patients is comparable to global figures. Future validation of the identified CNVs will involve long-read sequencing and exome sequencing will be conducted in patients with single PRKN deletions. This study sheds light on the genetic landscape of PD in Kazakhstan, facilitating further research and therapeutic interventions.
Table 1
To cite this abstract in AMA style:
R. Kaiyrzhanov, N. Zharkynbekova, S. Abdraimova, C. Shashkin, Z. Myrzayev, A. Karimova, V. Akhmetzhanov, K. Mok, J. Hardy, H. Houlden. Investigating Copy Number Variations in SNCA and PRKN in a Cohort of Kazakhstani Patients with Parkinson’s Disease and Healthy Controls [abstract]. Mov Disord. 2024; 39 (suppl 1). https://www.mdsabstracts.org/abstract/investigating-copy-number-variations-in-snca-and-prkn-in-a-cohort-of-kazakhstani-patients-with-parkinsons-disease-and-healthy-controls/. Accessed January 4, 2026.« Back to 2024 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/investigating-copy-number-variations-in-snca-and-prkn-in-a-cohort-of-kazakhstani-patients-with-parkinsons-disease-and-healthy-controls/