Category: Parkinson's Disease: Genetics
Objective: The objectives of the current study were to explore how the fecal metabolomics changed, and how they interacted with the alteration of microbiota in patients with Parkinson’s disease (PD). The reliable biomarkers for PD patients based on this multi-omics analysis was also aimed to identify.
Background: Cumulative evidences suggested that gut microbial disturbances were implicated in PD. How gut microbiota composition and fecal metabolites relate to PD is largely unknown. Up to date, there have been no report about the association between fecal metabolome and metagenome in a Chinese cohort of PD patients.
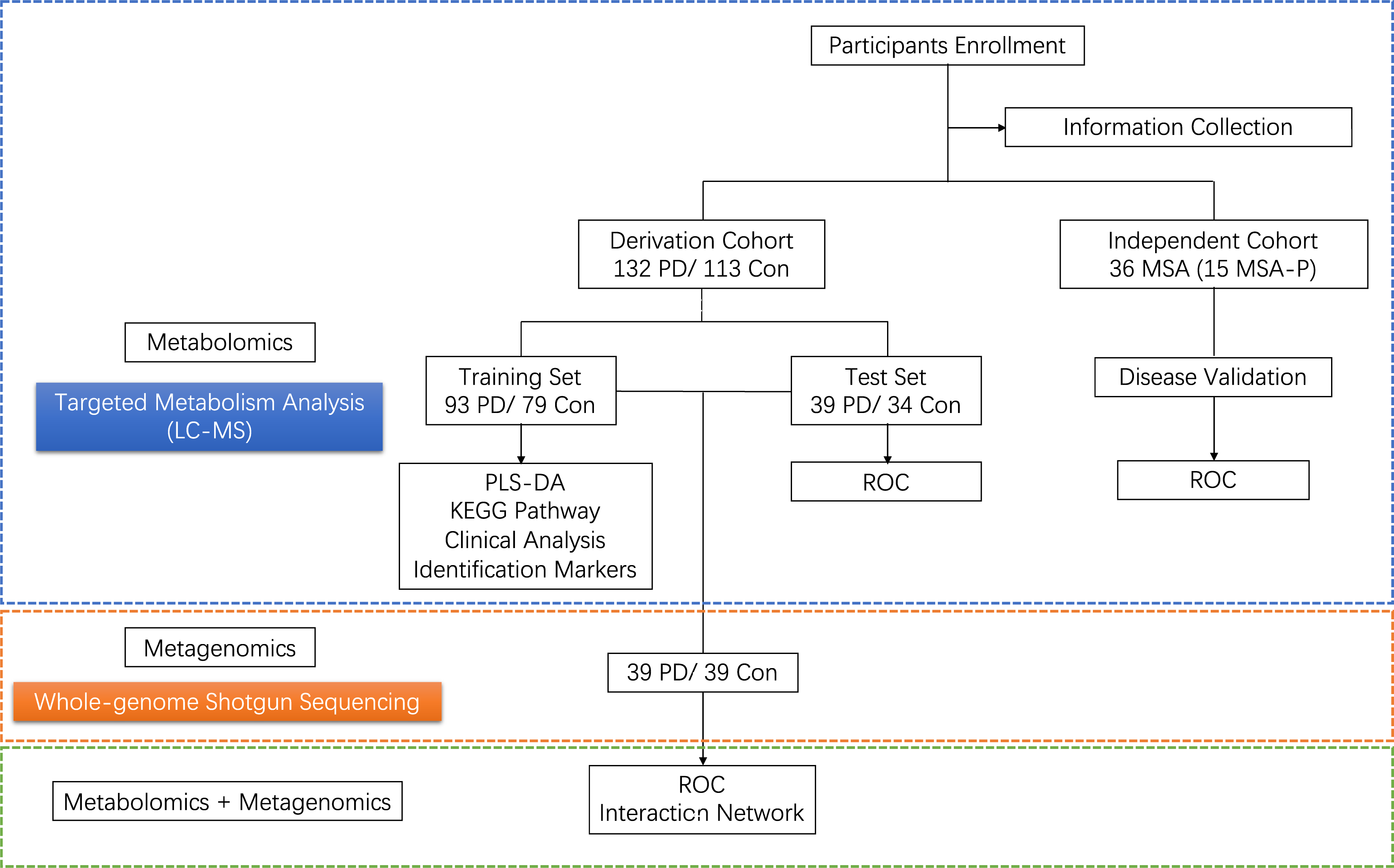
Method: In this study, targeted metabolomics was performed on the fecal samples of 132 patients with PDs, 113 healthy controls (HCs) and 36 patients with multiple system atrophy (MSA) using liquid chromatography-tandem mass spectrometry (LC-MS) method. The data of shotgun metagenome was analyzed among 39 PD patients and 39 HCs. By integrating metabolomics and metagenomic data, we characterized the connection between fecal metabolomics alterations and gut microbiota disturbance.
Results: A total of 33 disturbed fecal metabolites were identified between PD patients and HCs. A panel of 12 fecal metabolites were of greatest importance to distinguish PD patients from HCs, as well as from MSA patients. Using multi-omics data, we outlined the interaction networks of differential microbial species, microbial genes and fecal metabolites and we found the disturbed microbial genes and fecal metabolites were mainly mapped to amino acid metabolism. In addition, we also discovered a combinatorial panel of the fecal metabolites and microbial gene markers that could distinguish PD from HCs with high accuracy.
Conclusion: Taken together, we observed metabolomics dysregulation in fecal samples of PD patients. We also identified the relationships between the features of the gut microbiota and fecal metabolites, providing new clues for understanding the roles of the overall gut ecosystem in PD pathogenesis and may facilitate developing novel diagnostic strategies for PD.
References: 1. Qian, Y., et al. Gut metagenomics-derived genes as potential biomarkers of Parkinson’s disease. Brain 143, 2474-2489 (2020).
2. Jeffery, I.B., et al. Differences in Fecal Microbiomes and Metabolomes of People With vs Without Irritable Bowel Syndrome and Bile Acid Malabsorption. Gastroenterology 158, 1016-1028 e1018 (2020).
To cite this abstract in AMA style:
Y. Qian, S. Xu, X. He, Y. Lai, Y. Zhang, C. Mo, P. Ai, X. Yang, Q. Xiao. Integrative analysis of fecal metabolomics and metagenomics for Parkinson’s disease [abstract]. Mov Disord. 2023; 38 (suppl 1). https://www.mdsabstracts.org/abstract/integrative-analysis-of-fecal-metabolomics-and-metagenomics-for-parkinsons-disease/. Accessed December 25, 2025.« Back to 2023 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/integrative-analysis-of-fecal-metabolomics-and-metagenomics-for-parkinsons-disease/