Objective: To identify replicated metabolic markers of Parkinson’s Disease (PD) diagnosis from metabolomic studies and predict additional metabolic markers using computational metabolic modelling.
Background: Reported metabolic markers of PD are highly heterogeneous, making it difficult to conclude the role of metabolism in PD.
Method: The metabolites associated with a diagnosis of PD were compiled from published metabolomic studies. The consistency of these metabolic markers was explored. Replicated metabolites that were reported in at least two studies with the same changing trends were considered to be reliable biomarkers. To explore dysfunctional pathways of PD, all the metabolites were cross-matched with the metabolites in a computational model of human metabolism (Recon3D). Manual reconstruction was used to extend this global model with additional metabolic pathways of PD. Subsequently, an established pipeline (XomicsToModel) was used to generate a PD metabolic model with a minimal set of pathways linking the replicated metabolites.
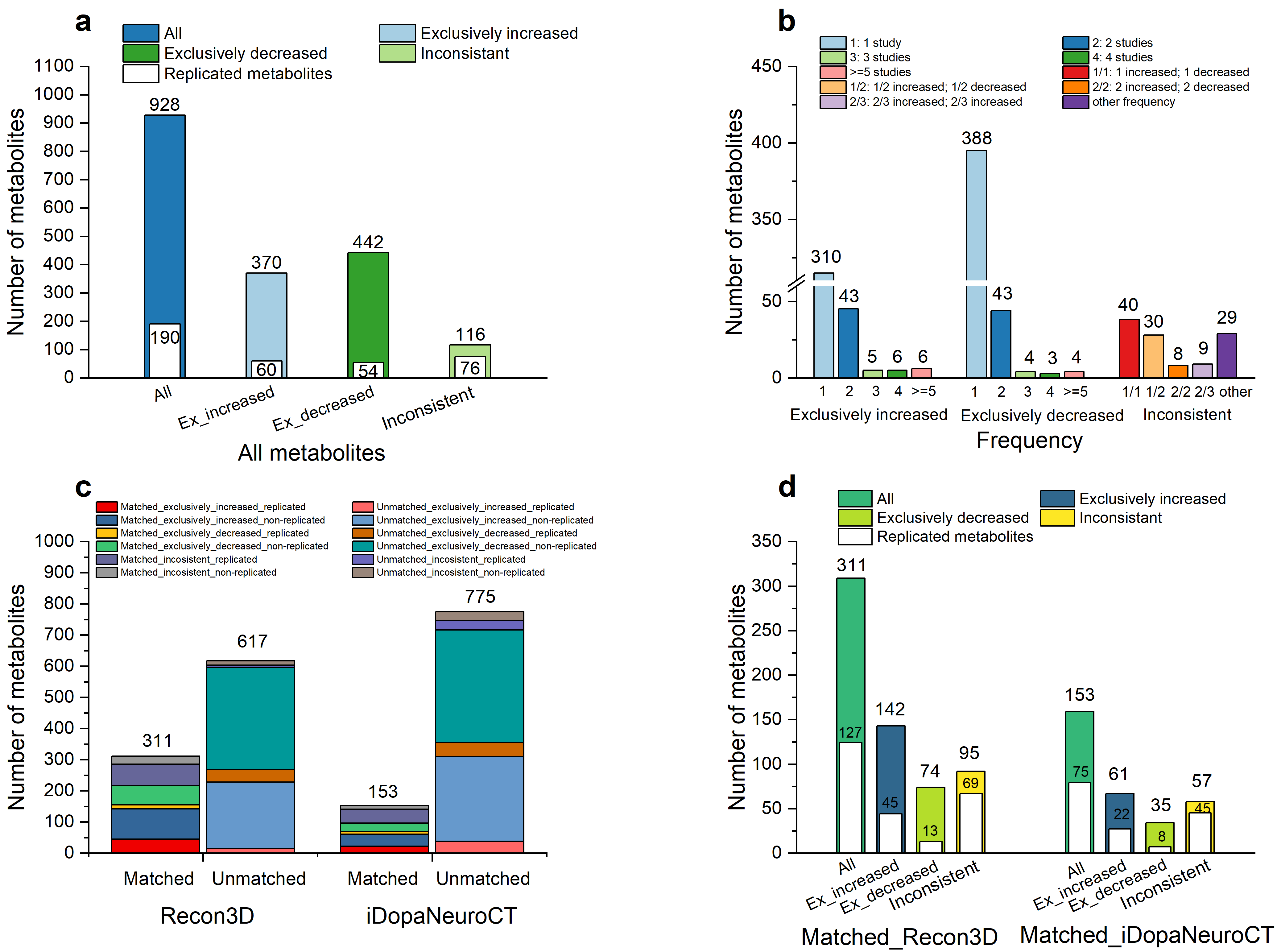
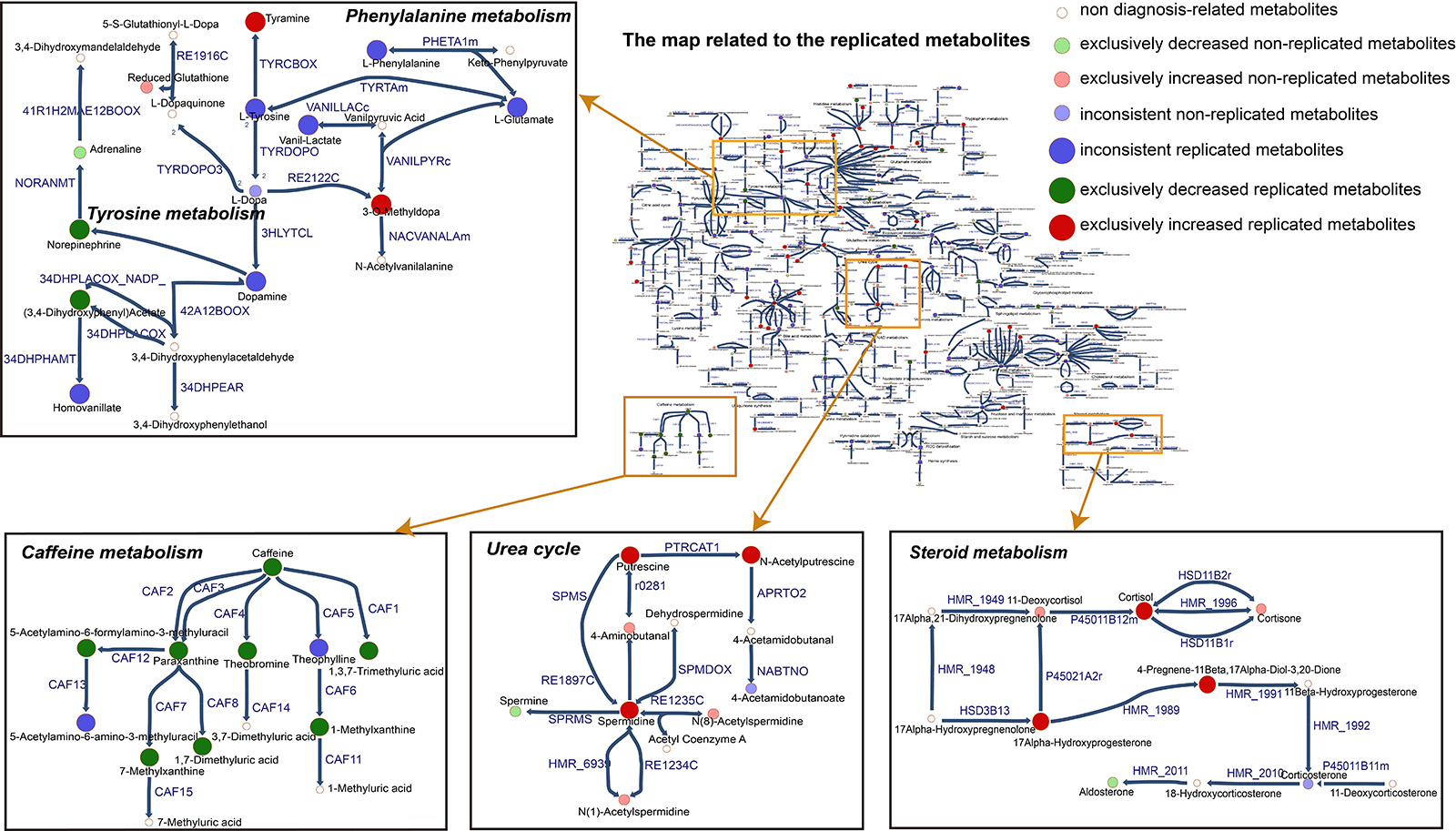
Results: 928 diagnosis-associated metabolites were collected from 74 studies, with 370 metabolites exclusively increased, 442 metabolites exclusively decreased, and 116 metabolites inconsistently changed in different studies. The 311 metabolites matched with Recon3D included 127 replicated metabolites [figure1]. Perturbation of caffeine metabolism was identified in PD patients. The refined global model contained all reactions from Recon3D, plus 277 new reactions from a complementary metabolic model (Human1), 14 from an established dopaminergic neuronal metabolic model, 73 novel fatty acid oxidation reactions and 43 caffeine metabolism reactions. A total of 137 replicated metabolites were used to extract a PD metabolic model that contained 910 metabolites, 1,689 reactions and 808 metabolic genes. Several metabolites, including cortisol and its metabolites, dopamine metabolites, polyamines, and caffeine metabolites, seem to be potential metabolic markers of PD [figure2].
Conclusion: A comprehensive meta-analysis of metabolomic studies identified 137 metabolites associated with PD. Corresponding metabolic pathways and their relationships were reviewed in detail. Computational modelling of PD metabolism enables an enhanced understanding of dysfunctional pathways in PD and the prediction of possible markers.
References: [1] E. Brunk et al. “Recon3D: A Resource Enabling A Three-Dimensional View of Gene Variation in Human Metabolism”. In: Nature biotechnology 36.3 (Mar. 2018), pp. 272–281.
[2] L. Heirendt et al. “Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0”. eng. In: Nature Protocols 14.3 (Mar. 2019), pp. 639–702.
[3] G. Preciat et al. “Mechanistic model-driven exometabolomic characterisation of human dopaminergic neuronal metabolism”. In: bioRxiv (Jan. 2021), p. 2021.06.30.450562.
[4] G. Preciat et al. XomicsToModel: Multiomics data integration and generation of thermodynamically consistent metabolic models. en. Tech. rep. Company: Cold Spring Harbor Laboratory Distributor: Cold Spring Harbor Laboratory Label: Cold Spring Harbor Laboratory Section: New Results Type: article. Nov. 2021, p. 2021.11.08.467803.
To cite this abstract in AMA style:
X. Luo, YJ. Liu, A. Balck, C. Klein, R. Fleming. Identification of reproducible metabolites for Parkinson’s Disease diagnosis using clinical data and computational modelling [abstract]. Mov Disord. 2023; 38 (suppl 1). https://www.mdsabstracts.org/abstract/identification-of-reproducible-metabolites-for-parkinsons-disease-diagnosis-using-clinical-data-and-computational-modelling/. Accessed October 21, 2025.« Back to 2023 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/identification-of-reproducible-metabolites-for-parkinsons-disease-diagnosis-using-clinical-data-and-computational-modelling/