Objective: To prospectively investigate the association between plasma microbial metabolites and Parkinson’s disease (PD) risk within a metabolomics framework.
Background: Gut microbiota is altered in individuals with PD (1). Metabolites derived from microbiota activity have been utilized as functional indicators to investigate the gut-brain interaction in PD by exploratory case-control studies (2, 3). However, these studies were typically small-scale and adopted post-diagnosis measurements, which are susceptible to reverse causality.
Method: A nested case-control study was conducted within the EPIC prospective population cohort, comprising 351 cases with incident PD and 351 controls who were matched on age, sex and study center. Plasma samples were obtained at the cohort recruitment, on average 8 years (ranged 0.2-18 years) before PD diagnosis for the cases. The untargeted metabolome was profiled in plasma, with 167 microbial metabolites annotated. We performed a metabolome-wide association study (MWAS) to identify metabolites associated with future PD risk. Microbiota-relevant pathways related to PD risk were explored across different groups based on sex, smoking status, body mass index, and lipopolysaccharide-binding protein (LBP) levels as a biomarker of intestinal permeability (4).
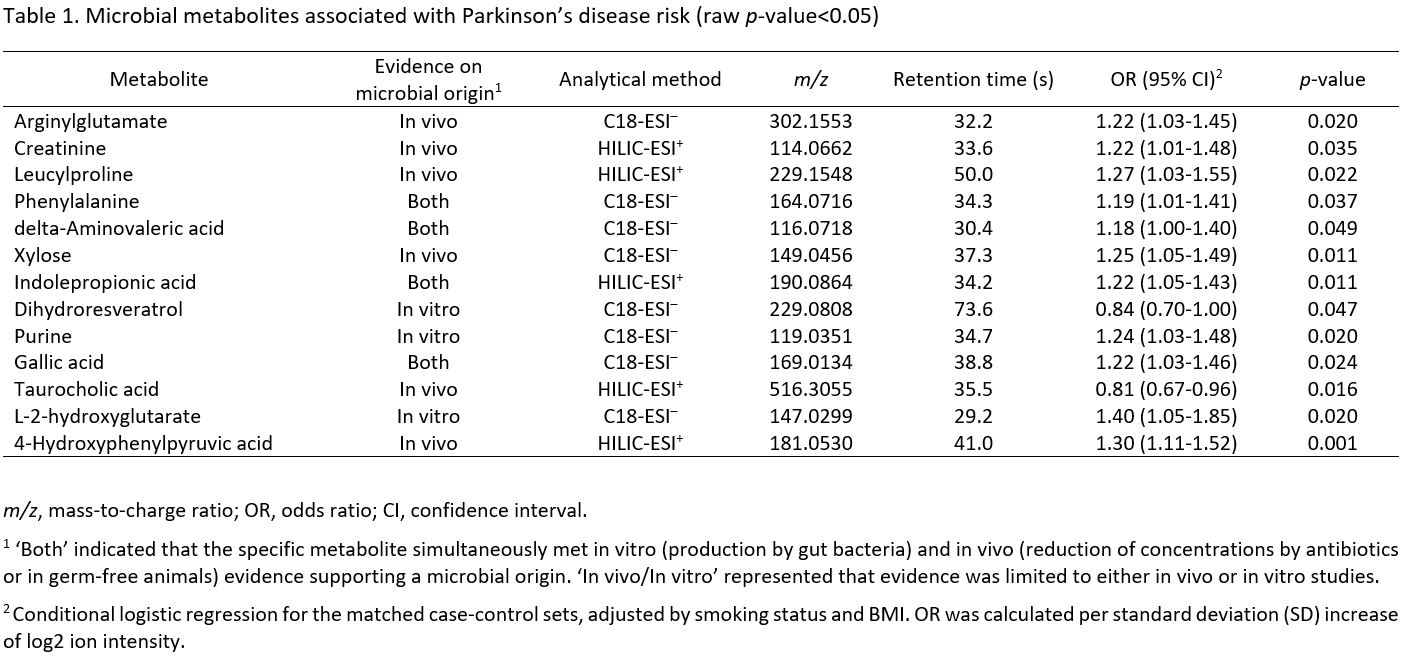
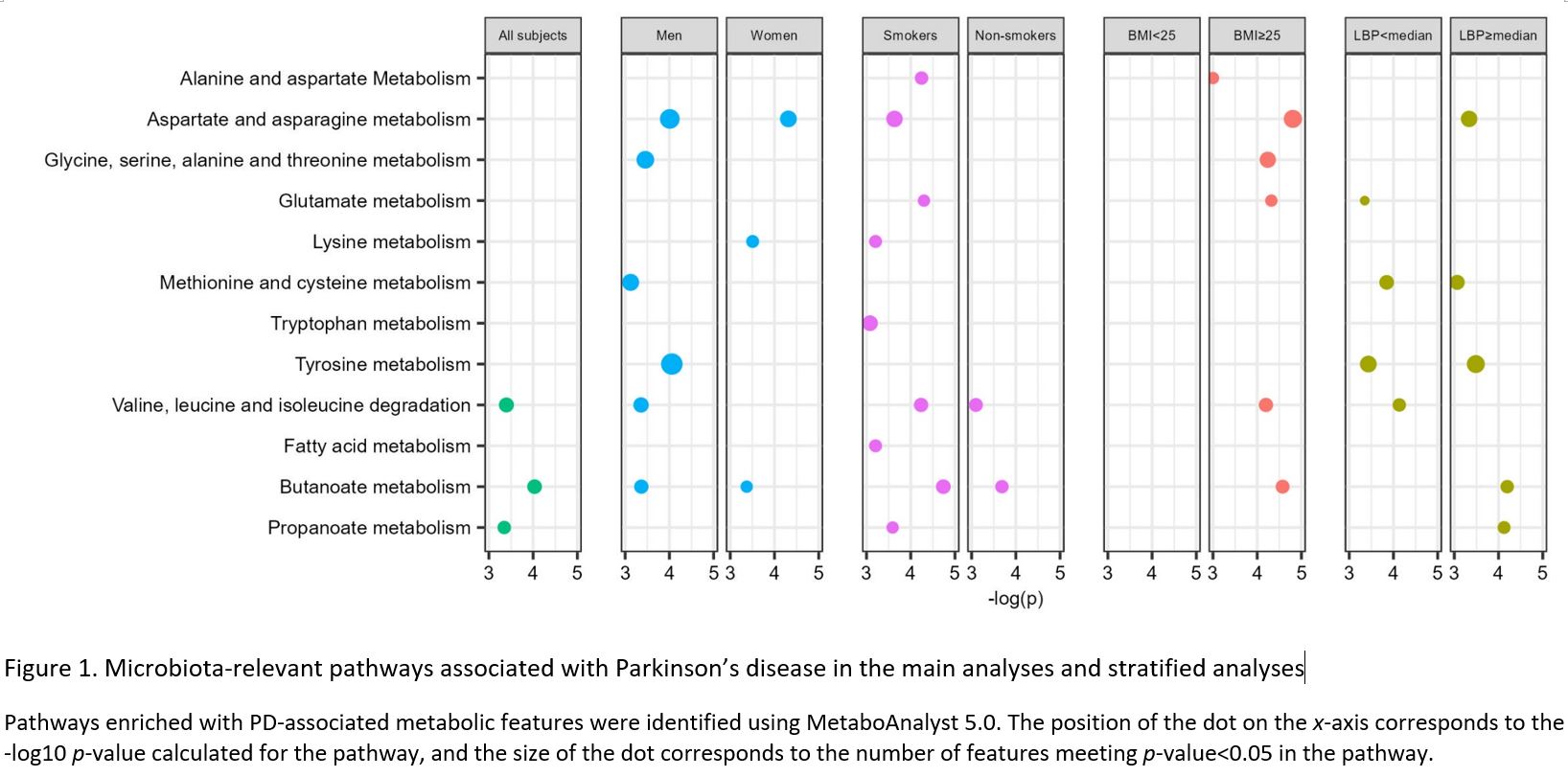
Results: Our MWAS identified 13 microbial metabolites which were nominally associated with PD risk (p-value<0.05), including amino acids, bile acid, indoles, and hydroxy acid (Table 1). However, none of these results remained significant after false discovery control (FDR=20%). Pathway analyses across the whole metabolome implicated three pathways in PD risk: (i) valine, leucine and isoleucine degradation, (ii) butanoate metabolism, and (iii) propanoate metabolism. In stratified analyses, more pronounced PD-associated microbial pathways were found in men, smokers, overweight/obese individuals, and those with higher LBP levels (Figure 1).
Conclusion: This study suggests that changes in microbial metabolites may be a pre-diagnostic feature of PD. We observed biologically plausible associations between microbial pathways and PD, and our results indicate that pathway enrichment may depend on individual characteristics.
Table 1
Figure 1
References: 1. Tan AH, Lim SY, Lang AE. The microbiome-gut-brain axis in Parkinson disease – from basic research to the clinic. Nat Rev Neurol. 2022;18(8):476-95.
2. Shao Y, Li T, Liu Z, et al. Comprehensive metabolic profiling of Parkinson’s disease by liquid chromatography-mass spectrometry. Mol Neurodegener. 2021;16(1):4.
3. Chen SJ, Chen CC, Liao HY, et al. Association of Fecal and Plasma Levels of Short-Chain Fatty Acids With Gut Microbiota and Clinical Severity in Patients With Parkinson Disease. Neurology. 2022;98(8):e848-e58.
4. Zhao Y, Walker DI, Lill CM, et al. Lipopolysaccharide-binding protein and future Parkinson’s disease risk: a European prospective cohort. J Neuroinflammation. 2023;20(1):170.
To cite this abstract in AMA style:
Y. Zhao, Y. Lai, S. Darweesh, B. Bloem, L. Forsgren, J. Hansen, JM. Huerta, M. Guevara, C. Lill, G. Miller, S. Peters, R. Vermeulen. Gut Microbial Metabolites and Future Risk of Parkinson’s Disease: A Metabolome-wide Association Study [abstract]. Mov Disord. 2024; 39 (suppl 1). https://www.mdsabstracts.org/abstract/gut-microbial-metabolites-and-future-risk-of-parkinsons-disease-a-metabolome-wide-association-study/. Accessed December 22, 2025.« Back to 2024 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/gut-microbial-metabolites-and-future-risk-of-parkinsons-disease-a-metabolome-wide-association-study/