Category: Parkinson's Disease: Neuroimaging
Objective: To investigate the influence of APOE ε4 allele on metabolic network activity and functional connectivity in patients with Parkinson’s disease (PD).
Background: Disease progression in PD is known to be influenced by genetic risk factors. The APOE ε4 allele represents a well-established risk factor for sporadic Alzheimer’s disease (AD). Studies, however, show conflicting results whether the APOE ε4 variant does influence motor and cognitive decline in PD. Recent preclinical evidence indicates that APOE ε4 regulates α-synuclein pathology, but the neural substrates of APOE ε4 in PD remain poorly understood.
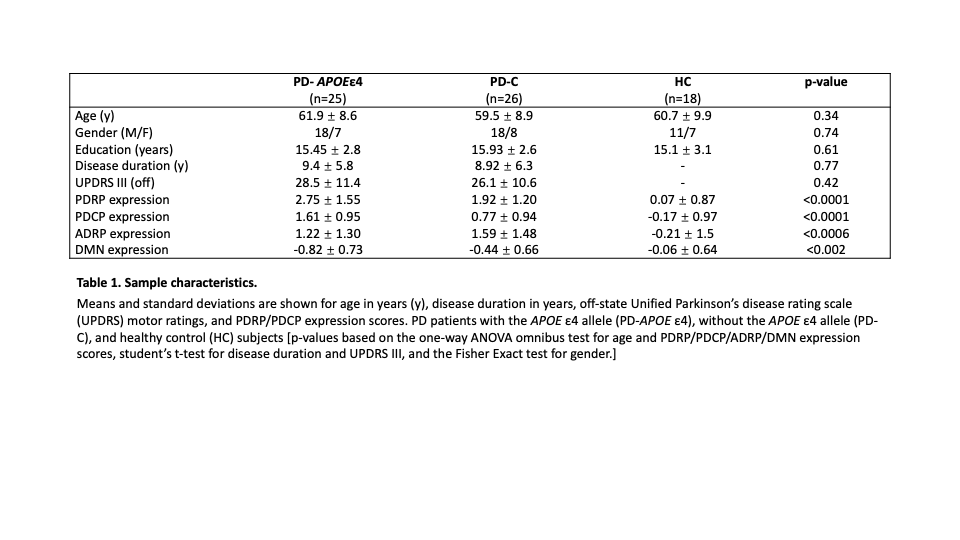
Method: We studied groups of PD patients with the APOE ε4 allele (PD-APOε4, n=25), without the APOE ε4 allele (PD-C, n=26), and 18 healthy controls (HC), that were matched for disease severity, duration, and demographics (Table 1). All participants were non-demented and underwent resting state imaging with [18F]-fluorodeoxyglucose (FDG) PET. SPM software was used to compare groups using a statistical threshold of p<0.01. Expression values of PD and AD related functional networks (PDRP and ADRP) and cognition related networks (PD cognition related network, PDCP and Default Mode Network, DMN) were computed in all subjects using an automated algorithm blind to genotypic classification. We also used graph theory to assess connectivity differences in network parameters within the different network spaces [2].
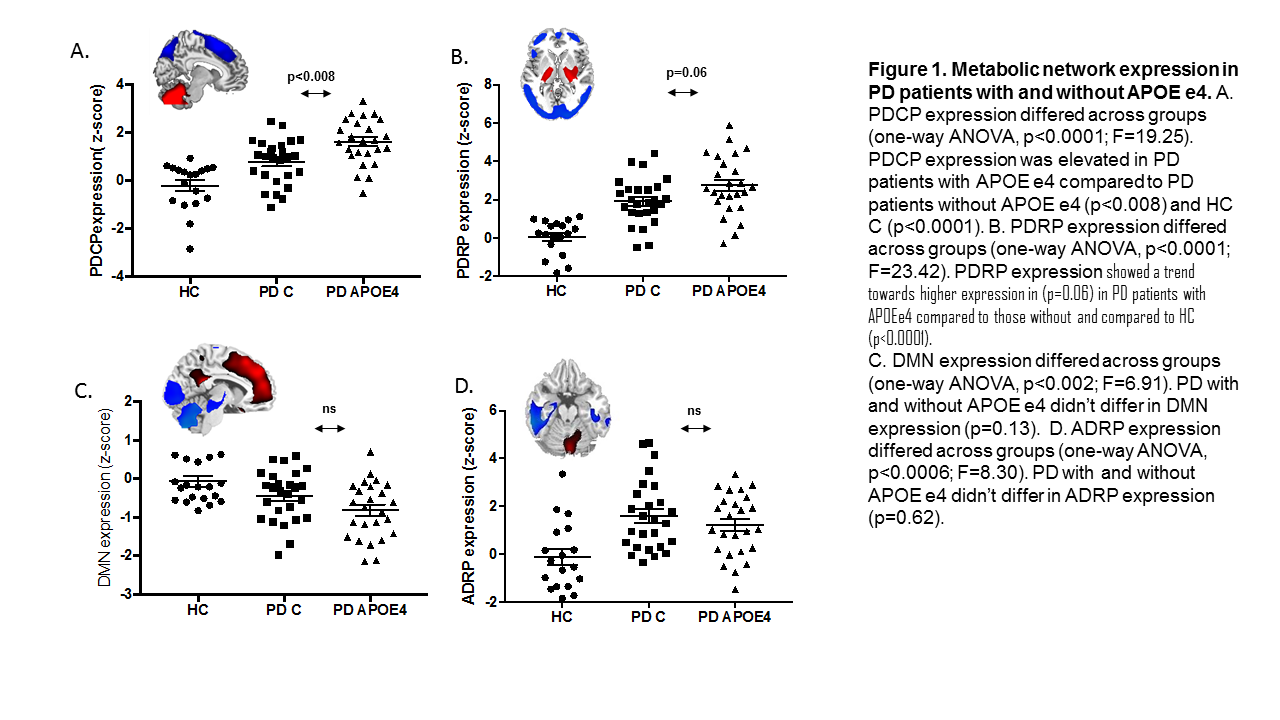
Results: Compared to PD-C, the PD-APOE ε4 group had reduced glucose metabolism in the superior frontal gyrus bilateral (p<0.02). PD patients with APOE ε4 exhibited abnormal increases in the PDCP network (p<0.008) compared to those without APOE ε4 (Figure 1). The motor related network showed a trend towards higher expression in (p=0.06) in PD patients with APOE ε4 allele compared to those without. ADRP and DMN expression values didn’t differ across groups (p>0.13). The PD-APOE ε4 group exhibited abnormal increases in network connectivity that were not present in the PD-C group.
Conclusion: PD patients carrying the unfavorable APOE ε4 allele exhibit reduced glucose metabolism in cortical superior frontal brain regions. We also found that this group had elevated PDCP expression levels compared to their duration- and severity-matched PD control counterparts. Hence, respective changes are related to PD specific networks and not mediated through changes in DMN or AD related networks.
References: [1] Schindlbeck KA, Eidelberg D. Network imaging biomarkers: insights and clinical applications in Parkinson’s disease. Lancet Neurol 2018
[2] Schindlbeck KA, Vo A, Nguyen N, Tang CC, Niethammer M, Dhawan V, Brandt V, Saunders-Pullman R, Bressman SB, Eidelberg D. LRRK2 and GBA Variants Exert Distinct Influences on Parkinson’s Disease-Specific Metabolic Networks. Cereb Cortex 2020
To cite this abstract in AMA style:
K. Schindlbeck, A. Vo, P. Mattis, N. Nguyen, D. Eidelberg. Functional network correlates of APOE ε4 allele in Parkinson’s disease [abstract]. Mov Disord. 2023; 38 (suppl 1). https://www.mdsabstracts.org/abstract/functional-network-correlates-of-apoe-%ce%b54-allele-in-parkinsons-disease/. Accessed October 17, 2025.« Back to 2023 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/functional-network-correlates-of-apoe-%ce%b54-allele-in-parkinsons-disease/