Session Information
Date: Tuesday, September 24, 2019
Session Title: Environmental Causes
Session Time: 1:45pm-3:15pm
Location: Agora 2 West, Level 2
Objective: Assess gene-pesticide associations with PD using an exposed-only design in a highly exposed cohort.
Background: Pesticide use is associated with an increased risk of PD. Genetic modification of this risk is difficult to assess and has been inconsistently reported. Exposed-only study designs may increase resolution to detect genetic interactions.[1]
Method: We recruited people with PD (PwPD) and matched controls from the Agricultural Health Study, a cohort of pesticide applicators and their spouses. PD was determined by expert exam, and pesticide usage by interview. DNA was obtained from peripheral blood. Genotyping used the Affymetrix UK Biobank Array, with filtering of SNPs with call rates <95%, HWE p<10-6, and allele frequencies <1%. For the current analysis, we pre-specified 123 metabolic and antioxidant genes of interest, and conducted analyses within 5 subcohorts with exposures to specific pesticides: rotenone, Paraquat, permethrin, cyclodienes, and 2,4-D. We calculated risk for each of 3309 SNPs individually. We also used EPACTS toolbox (https://genome.sph.umich.edu/wiki/EPACTS) to implement SKAT-O gene-wise group burden tests to integrate the total SNP variability across each gene[2] adjusting for age, sex, smoking and state (NC or IA).
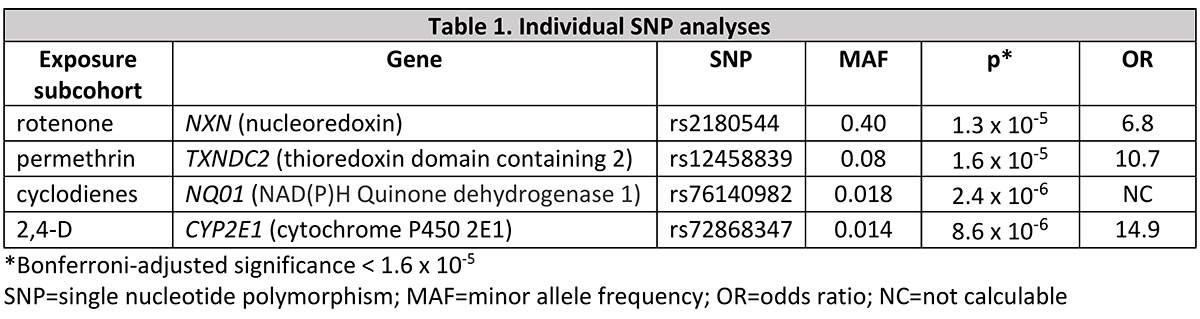
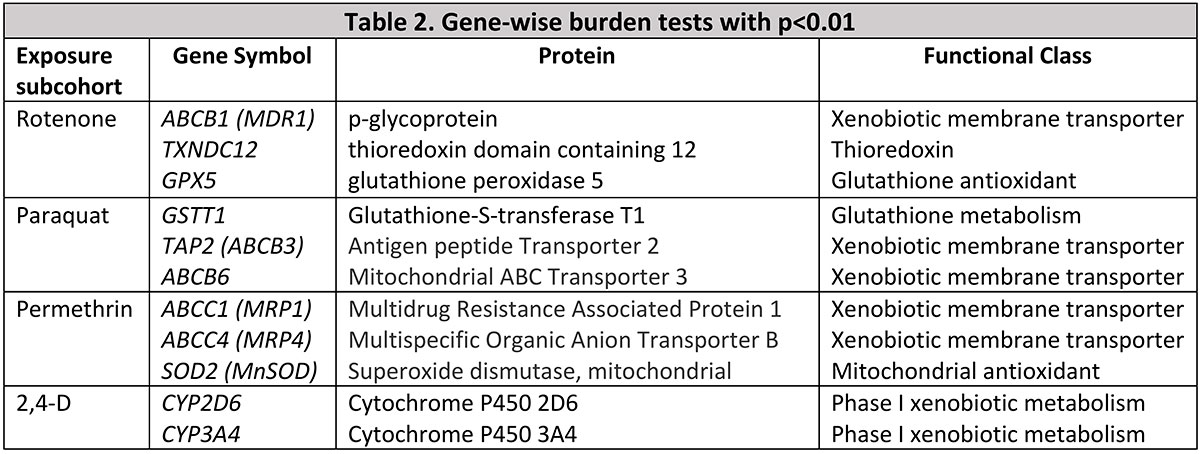
Results: DNA was available for 100 PwPD and 371 controls, including rotenone (19 PwPD, 31 control), Paraquat (22 PwPD, 52 control), permethrin (16 PwPD, 43 control), cyclodiene (30 PwPD, 109 control), and 2,4-D (72 PwPD, 254 control) exposed subcohorts. After adjustment for multiple comparisons, several individual SNPs were significantly associated with PD in the exposed subcohorts [table1]. Gene-burden tests were not significantly associated after false discovery rate adjustment, but several genes involved in pesticide metabolism and xenobiotic membrane transporters tended toward significance [table2].
Conclusion: Despite small sample sizes, we were able to find significant interactions with individual SNPs in pesticide metabolic and transporter genes using an exposed-only design. Gene-burden tests implicate a range of additional pesticide-related genes, among them ABCB1, which encodes the xenobiotic transporter p-glycoprotein and for which rotenone is a likely substrate.[3] Ongoing whole genome sequencing in this population will allow us to integrate substantial additional rare variant data to improve resolution using this novel approach to identifying gene-environment interaction.
References: [1] Zhao, L.P., et al., Deciphering Genome Environment Wide Interactions Using Exposed Subjects Only. Genet Epidemiol, 2015. 39(5): p. 334-46. [2] Lee, S., M.C. Wu, and X. Lin, Optimal tests for rare variant effects in sequencing association studies. Biostatistics, 2012. 13(4): p. 762-75. [3] Lacher, S.E., et al., P-Glycoprotein Transport of Neurotoxic Pesticides. J Pharmacol Exp Ther, 2015. 355(1): p. 99-107.
To cite this abstract in AMA style:
J. Kaye, L. Lima, C. Tanner, S. Finkbeiner, R. Swanson, S. Goldman. Exposed-Only Analysis of Gene-Pesticide Interaction in Parkinson’s Disease (PD) [abstract]. Mov Disord. 2019; 34 (suppl 2). https://www.mdsabstracts.org/abstract/exposed-only-analysis-of-gene-pesticide-interaction-in-parkinsons-disease-pd/. Accessed December 20, 2025.« Back to 2019 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/exposed-only-analysis-of-gene-pesticide-interaction-in-parkinsons-disease-pd/