Category: Parkinson's Disease: Genetics
Objective: We aimed to identify novel candidate autosomal recessive (AR) genes combining whole-exome sequencing (WES) and long-read sequencing data (LRS) in autosomal recessive Parkinson’s disease (AR-PD) families and sporadic early-onset PD patients of the Chinese mainland population.
Background: As a complex neurodegenerative disorder, PD has been identified with more than 20 genes with Mendelian monogenic inheritance [1]. About 60% of early-onset AR-PD cases have been confirmed to carry pathogenic or likely pathogenic variants [2], suggesting that other potential AR-PD candidate genes might not have yet been discovered. The evidence suggested that there were still novel AR-PD candidate genes that might not have yet been discovered.
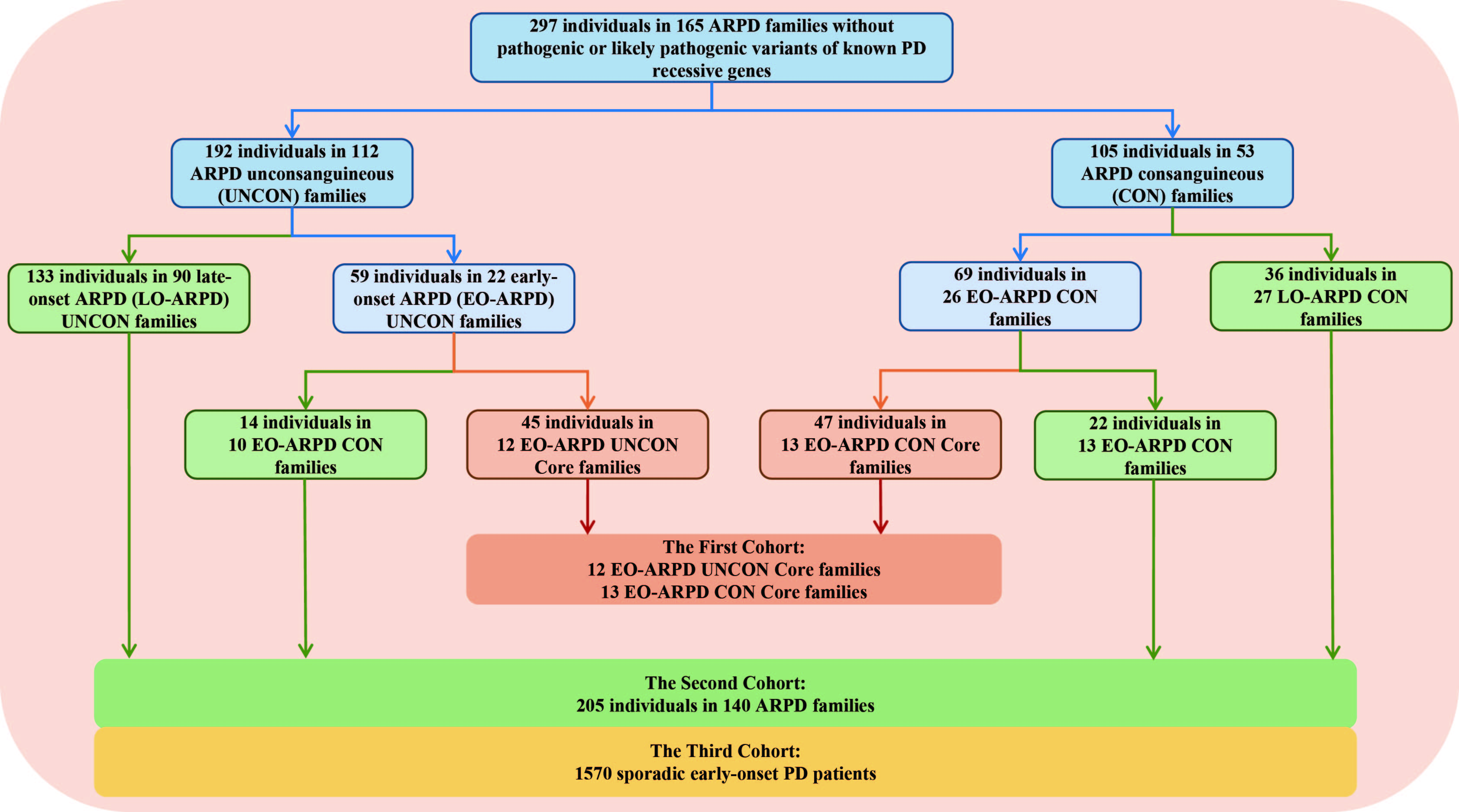
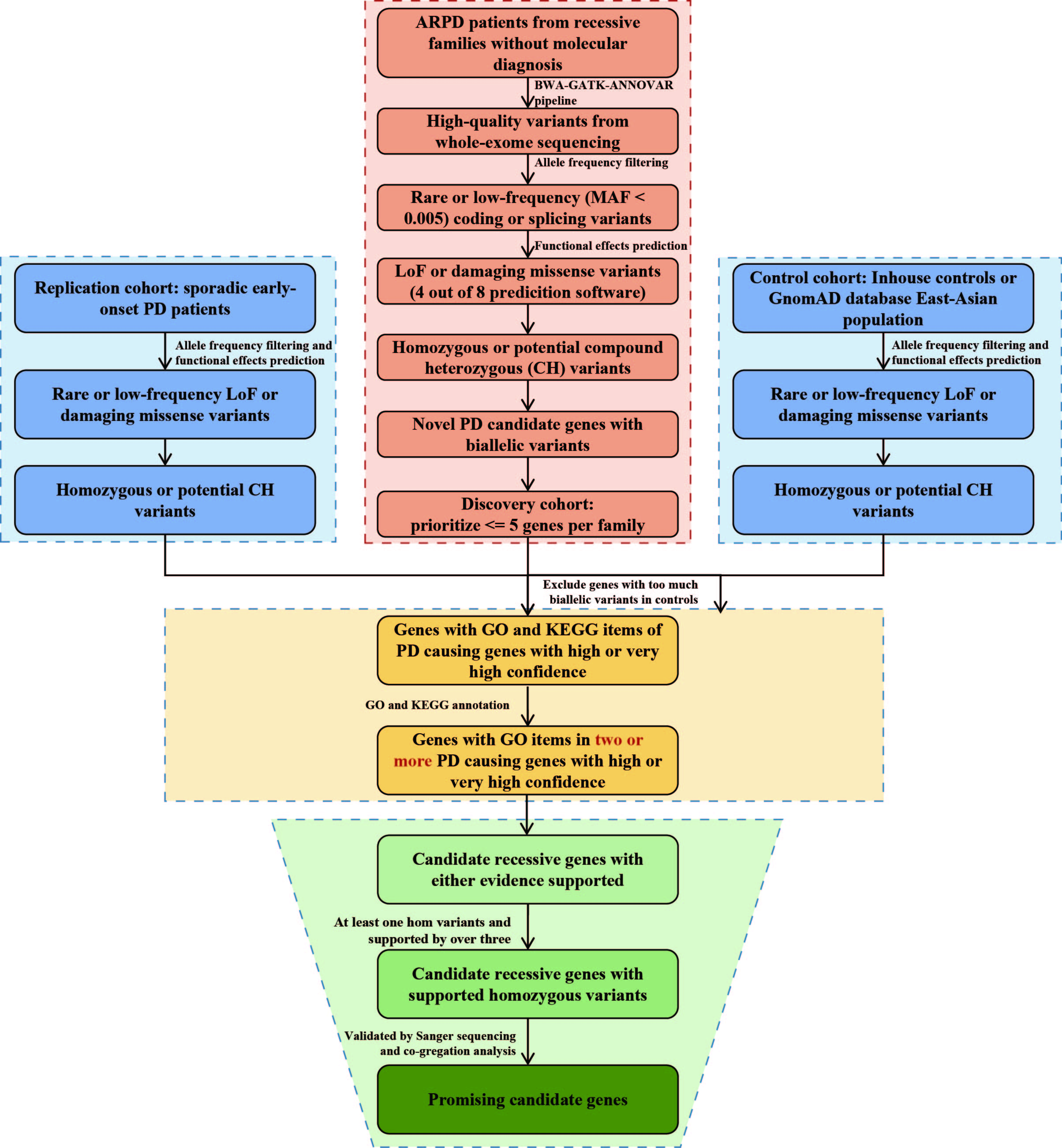
Method: A total of 162 AR-PD families and 1570 sporadic early-onset PD (sEOPD) patients from the the Parkinson’s Disease and Movement Disorders Multicenter Database and Collaborative Network in China (PD-MDCNC, http://pd-mdcnc.com/) database [3] were included in this study. The homozygous mapping regions were detected from the WES data by H3M2 [4] and HOMWES [5] software or methods, and then combined the biallelic variants identified from WES data to identify the candidate AR genes from the core AR-PD families. Next, we screened those candidate recessive genes in the replication sEOPD cohorts. (figure 1) Meanwhile, a bioinformatics analysis based on a population-based analysis of biallelic variants was conducted in this study. (figure 2) Furthermore, we analyzed LRS data to further explore candidate genes with complex variants.
Results: Based on the analysis of WES data from 25 core AR-PD families, we identified six candidate genes; combing the replication cohorts, we prioritized one promising gene. Based on the population-based prioritizing approach, we could identify eleven candidate genes and prioritize one promising gene. It is interesting to note that the two methods candidate is the same gene. More importantly, mutations at positions 275 and 332 lead to instability in this gene. Based on the analysis of LRS data from AR-PD families, we identified a candidate gene with a homozygous structure variant (6.3kb deletion).
Conclusion: Our study identified 17 candidate genes using a large sample of AR-PD families in the Chinese population, combining WES and LRS data, which may expand the spectrum of candidate autosomal recessive genes responsible for PD.
Flowchart of the enrolled patients
Flowchart of prioritizing approaches
References: [1]Zhao Y, Qin L, Pan H, et al. The role of genetics in Parkinson’s disease: a large cohort study in Chinese mainland population. Brain. 2020; 143: 2220-34.
[2]Blauwendraat C, Nalls MA, Singleton AB. The genetic architecture of Parkinson’s disease. Lancet Neurol. 2020; 19: 170-8.
[3]Zhou X, Liu Z, Zhou X, et al. The Chinese Parkinson’s Disease Registry (CPDR): Study Design and Baseline Patient Characteristics. Movement disorders : official journal of the Movement Disorder Society. 2022; 37: 1335-45.
[4]Magi A, Tattini L, Palombo F, et al. H3M2: detection of runs of homozygosity from whole-exome sequencing data. Bioinformatics. 2014; 30: 2852-9.
[5]Kancheva D, Atkinson D, De Rijk P, et al. Novel mutations in genes causing hereditary spastic paraplegia and Charcot-Marie-Tooth neuropathy identified by an optimized protocol for homozygosity mapping based on whole-exome sequencing. Genet Med. 2016; 18: 600-7.
To cite this abstract in AMA style:
Y. Zhao, H. Pan, J. Guo, Z. Liu, B. Tang. Expanding the Spectrum of Autosomal Recessive Genes Responsible for Parkinson’s Disease in the Chinese Population [abstract]. Mov Disord. 2024; 39 (suppl 1). https://www.mdsabstracts.org/abstract/expanding-the-spectrum-of-autosomal-recessive-genes-responsible-for-parkinsons-disease-in-the-chinese-population/. Accessed April 20, 2025.« Back to 2024 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/expanding-the-spectrum-of-autosomal-recessive-genes-responsible-for-parkinsons-disease-in-the-chinese-population/