Category: Parkinson's Disease: Neuroimaging
Objective: This study aimed using multimodal MRI to test two models of Parkinson’s disease propagation and investigated how the models were influenced by the presence of rapid eye movement sleep behavior disorder (RBD).
Background: The classical Braak neuropathological staging model in Parkinson’s disease (PD) suggests that brain lesions progress from the medulla oblongata to the cortex [1]. An alternative model in which neurodegeneration first occurs in the cortex has also been proposed [2-4]. These two models may correspond to different patient phenotypes, for instance, patients presenting with rapid eye movement sleep behavior disorder (RBD) or not [5-6].
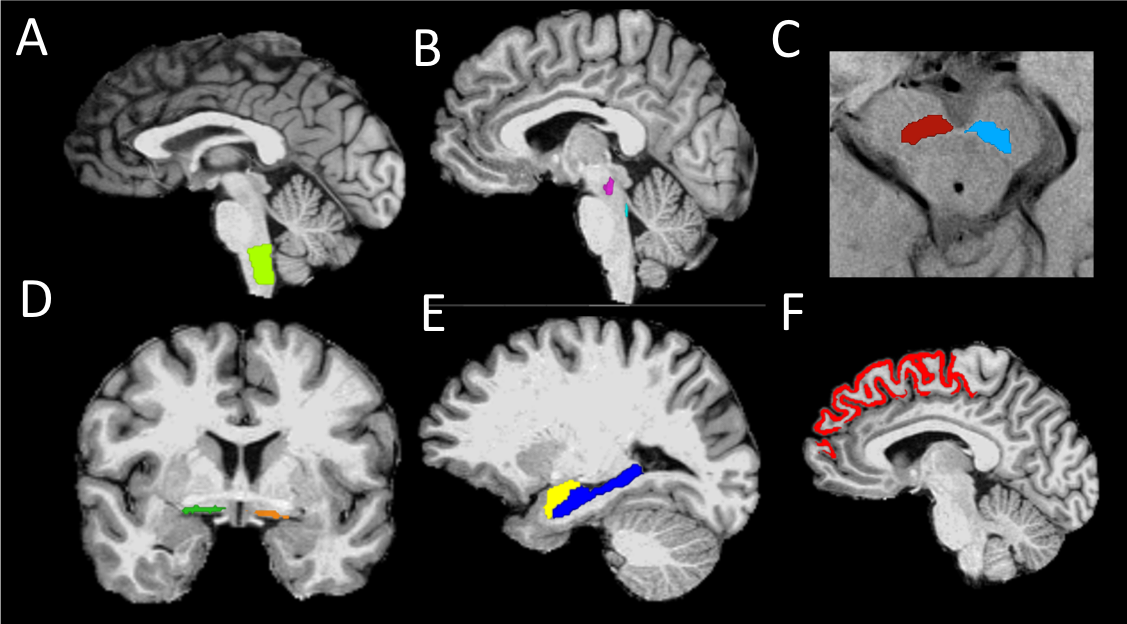
Method: Fifty-four patients with PD (thirty-four with RBD) and twenty-five healthy volunteers were scanned using T1-weighted, diffusion tensor and neuromelanin-sensitive imaging. Volume, signal, mean, axial and radial diffusivities were calculated in brainstem, basal forebrain and cortical regions (Figure 1).
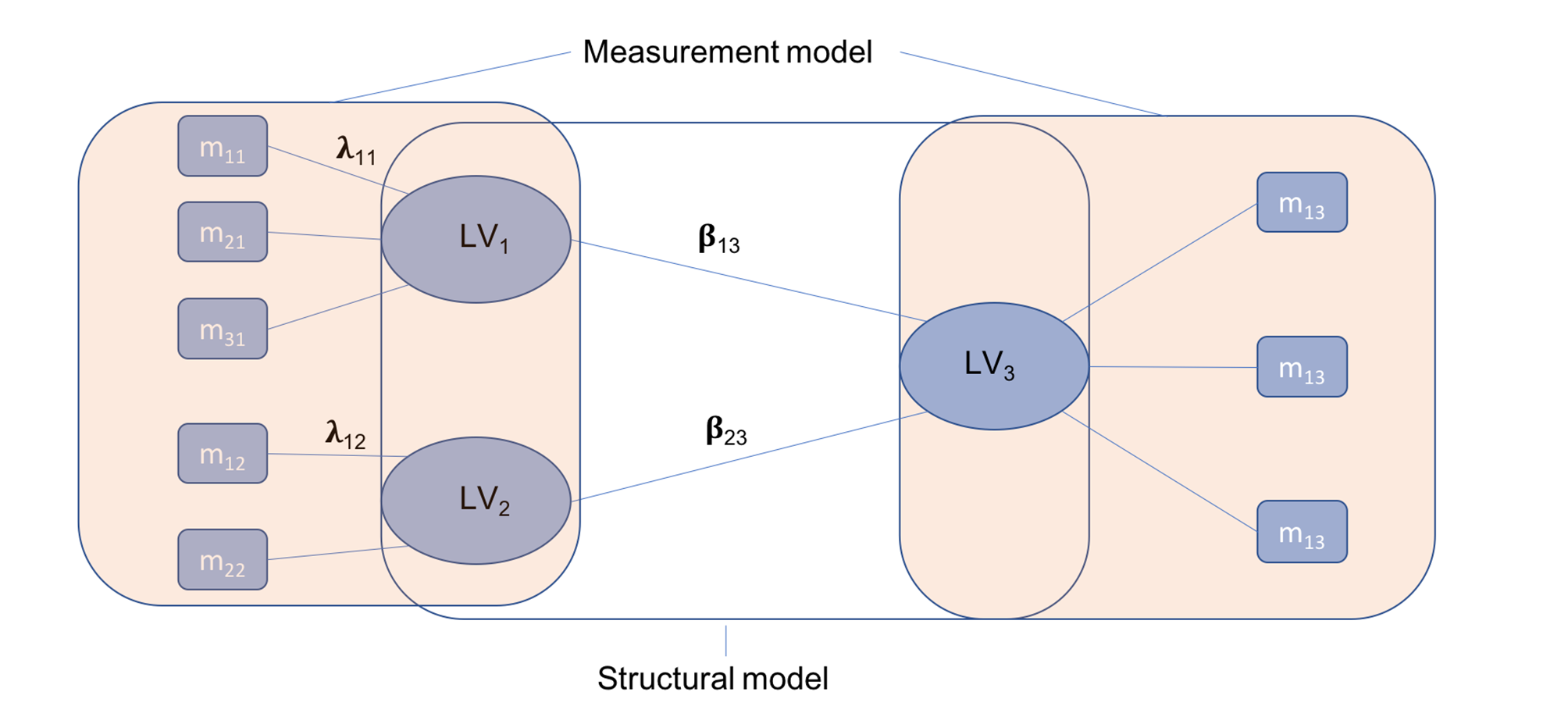
Partial Least Squares Path-Modeling (PLS-PM), estimating a network of causal relationships between blocks of variables was used to build and test an analytical model based on Braak staging. First, observed variables or manifest variables (MVs) were grouped into blocks or latent variables (LVs). Then, a three-step algorithm was run (Figure 2). As a result, a set of path coefficients and factor loadings were obtained. The overall quality of the model was assessed using the goodness-of-fit coefficient (Gof) [7].
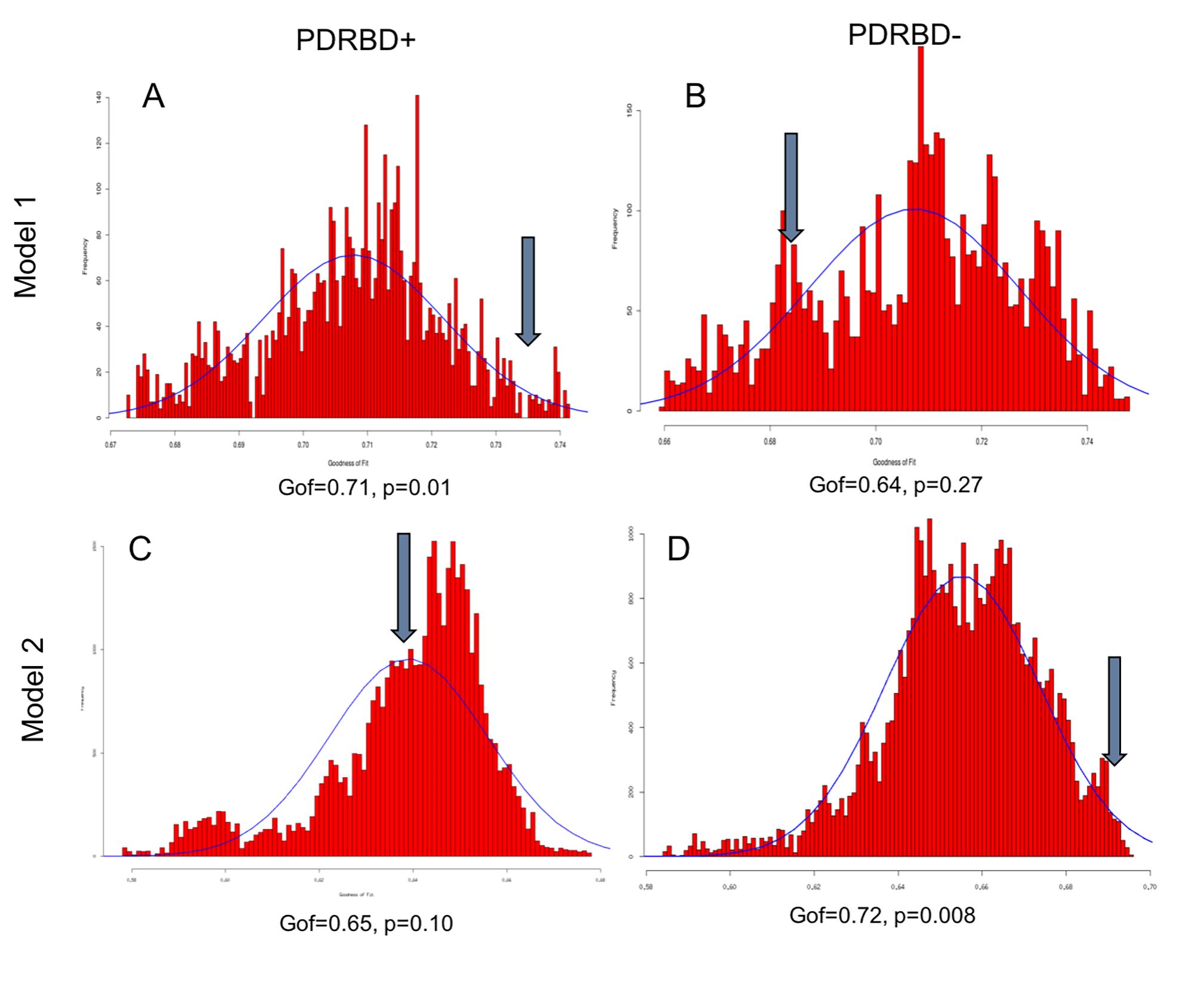
Results: PLS-PM was run on PD patients with RBD and without RBD separately (Figure 3). In PD with RBD, a brainstem-to-cortex model had significant Gof (0.71, p=0.01) whereas a cortex-to-brainstem model was not. In contrast, in PD patients without RBD, the brainstem-to-cortex model was not significant (Gof=0.64, p=0.27), and the cortex-to-brainstem model was highly significant (Gof=0.72, p=0.008).
Conclusion: Using the PLS-PM imaging-based model, the neurodegeneration pattern of PD patients with RBD (with greater damage in the brainstem) was consistent with the Braak brainstem-to-cortex model, whereas that of patients without RBD followed the cortex-to-brainstem model.
References: [1] Braak H. et al. Neurobiol Aging 2003;24:197–211. [2] Hawkes CH, Del Tredici K, Braak H. Neuropathol Appl Neurobiol. 2007;33:599–614. [3] Doty RL. Nat Rev Neurol. 2012;8:329–339. [4] Jellinger KA. Acta Neuropathol (Berl). 2008;116:1–16. [5] Knudsen K, Fedorova TD, Hansen AK, et al. Lancet Neurol. 2018;17:618–628. [6] Bauckneht M, Chincarini A, De Carli F, et al Sleep Med Rev. 2018;41:266–274. [7] Tenenhaus M, Amato S, Esposito Vinzi V. Proc XLII SIS Sci Meet. 2004;28:739–742.
To cite this abstract in AMA style:
N. Pyatigorskaya, L. Yahia-Cherif, R. Valabregue, R. Gaurav, F. Gargouri, C. Ewenczyk, C. Gallea, S. Fernandez-Vidal, I. Arnulf, M. Vidailhet, S. Lehericy. Dual hit model of Parkinson’s disease propagation using MRI biomarkers and partial least squares path modeling [abstract]. Mov Disord. 2020; 35 (suppl 1). https://www.mdsabstracts.org/abstract/dual-hit-model-of-parkinsons-disease-propagation-using-mri-biomarkers-and-partial-least-squares-path-modeling/. Accessed April 22, 2025.« Back to MDS Virtual Congress 2020
MDS Abstracts - https://www.mdsabstracts.org/abstract/dual-hit-model-of-parkinsons-disease-propagation-using-mri-biomarkers-and-partial-least-squares-path-modeling/