Category: Parkinson's Disease: Genetics
Objective: Here, we designed a case-control study to analyze WGS variants in NUS1 coding regions and NUS1 related non-coding regions to investigate the relative contribution of common and rare variants of NUS1 toward the development of sporadic late-onset Parkinson’s disease (LOPD) in the Han Chinese population.
Background: In our previous research, a two-stage sequencing-based strategy identified rare variants of the NUS1 gene in PD patients[1]. Moreover, RNAi NUS1 knockdown experiments in Drosophila induce PD-like phenotypes and abnormal cell apoptosis, thereby indicating that NUS1 plays an important role in the pathogenesis of PD. However, studies that have investigated the association of single genes with PD pathogenesis have largely focused on coding alterations in traditional drivers. To date, the association between the NUS1 gene and PD is seldom discussed.
Method: Using our whole genome sequencing including 1,962 cases and 1,279 controls to discover NUS1 coding and noncoding common and rare variants. Burden analysis was performed to evaluate rare variants’ association and LOPD. Logistic regression analysis was performed to evaluate common variants’ association and LOPD. Sequence Kernel Association test (SKAT) CommonRare function was used to evaluate for the role of the common and rare variants within noncoding regions in LOPD.
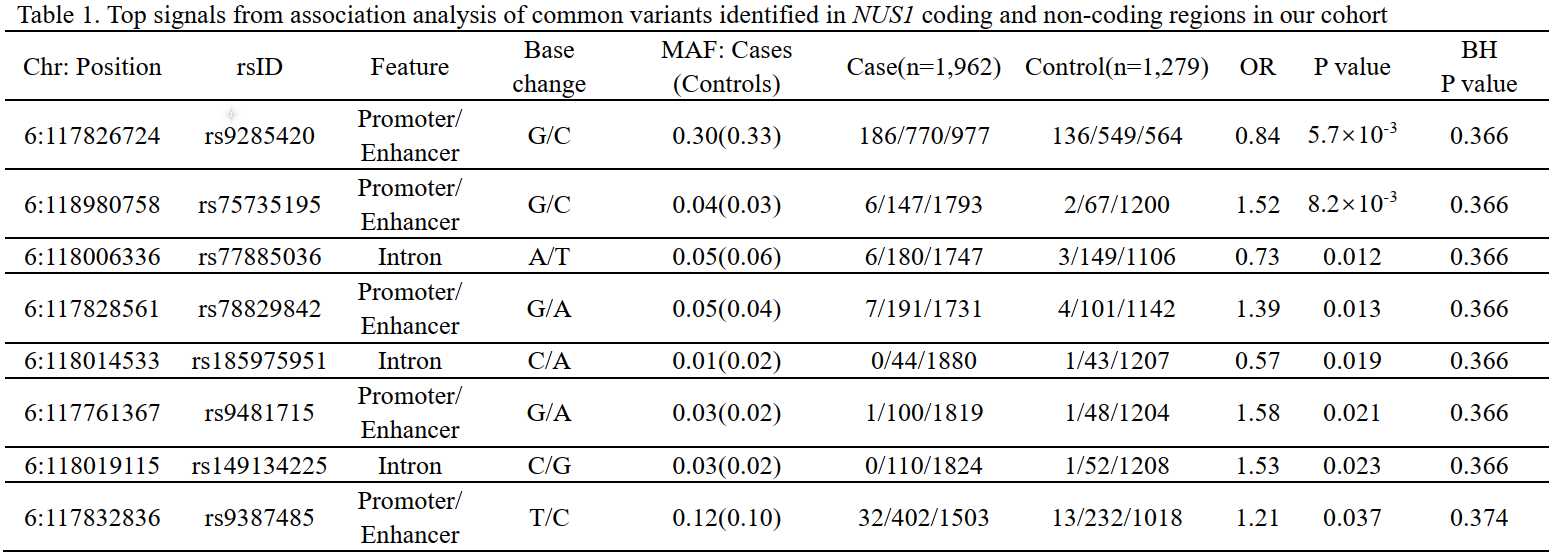
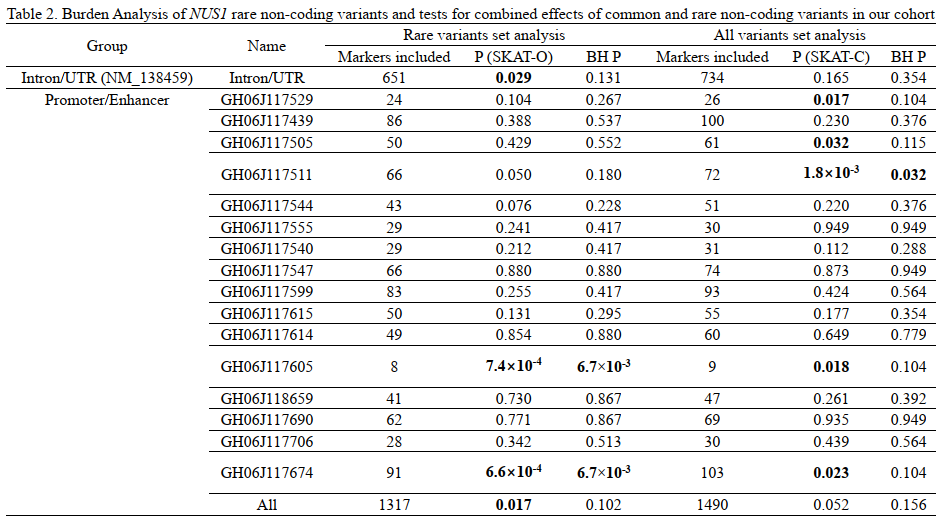
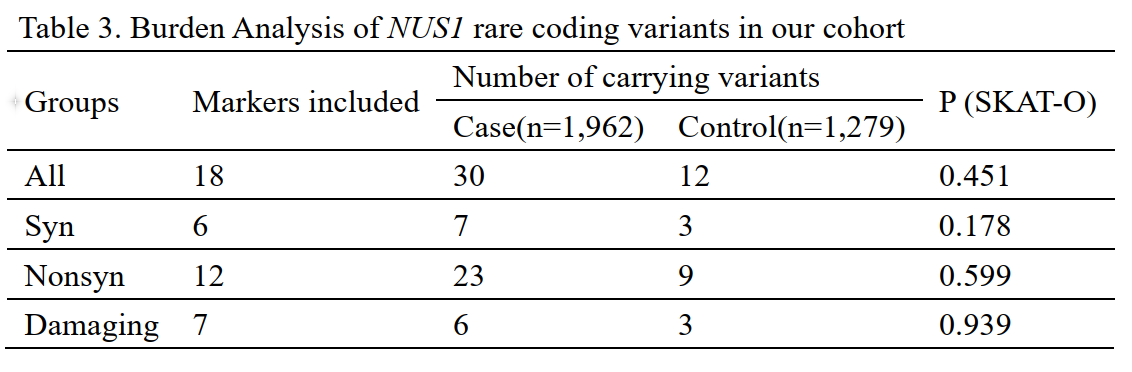
Results: Five index variantsrs75735195, rs78829842, rs9481715, rs149134225 and rs9387485) within the promoter/enhancer and intronic regions increased risk of LOPDOR>1; P<0.05), and three index variantsrs9285420, rs77885036 and rs185975951) within the and promoter/enhancer regions reduced the risk for LOPDOR<1; P<0.05[table 1]. However, none survived the correction for multiple testing. We found that GH06J117605 and GH06J117674 rare sets were associated with increased risks of LOPD after correction for multiple testingtable 2]. A significant aggregate contribution of rare and common variants in GH06J117511 to LOPD was found after correcting for multiple testing. No significant associations between coding variants of NUS1 and LOPD were foundtable 3].
Conclusion: These findings provide new information regarding the noncoding regions of the NUS1 in Chinese LOPD cohorts and indicate that rare variants or combined with common variants in of NUS1 contribute to the risk of LOPD.
References: [1] Guo JF, Zhang L, Li K, Mei JP, Xue J, Chen J, Tang X, Shen L, Jiang H, Chen C, Guo H, Wu XL, Sun SL, Xu Q, Sun QY, Chan P, Shang HF, Wang T, Zhao GH, Liu JY, Xie XF, Jiang YQ, Liu ZH, Zhao YW, Zhu ZB, Li JD, Hu ZM, Yan XX, Fang XD, Wang GH, Zhang FY, Xia K, Liu CY, Zhu XW, Yue ZY, Li SC, Cai HB, Zhang ZH, Duan RH, Tang BS. Coding mutations in NUS1 contribute to Parkinson’s disease. Proc Natl Acad Sci U S A 2018; 115:11567-72.
To cite this abstract in AMA style:
L. Jiang, H. Pan, Y. Zhao, Q. Zeng, Z. Liu, Q. Sun, Q. Xu, J. Tan, X. Yan, J. Li, B. Tang, J. Guo. Contribution of coding/noncoding variants in NUS1 to late-onset sporadic Parkinson’s disease [abstract]. Mov Disord. 2020; 35 (suppl 1). https://www.mdsabstracts.org/abstract/contribution-of-coding-noncoding-variants-in-nus1-to-late-onset-sporadic-parkinsons-disease/. Accessed December 19, 2025.« Back to MDS Virtual Congress 2020
MDS Abstracts - https://www.mdsabstracts.org/abstract/contribution-of-coding-noncoding-variants-in-nus1-to-late-onset-sporadic-parkinsons-disease/