Session Information
Date: Monday, October 8, 2018
Session Title: Parkinson's Disease: Neuroimaging And Neurophysiology
Session Time: 1:15pm-2:45pm
Location: Hall 3FG
Objective: To investigate whether the integration of PET imaging and metabolomics data can provide improved machine learning models for PD diagnosis.
Background: The reliable diagnosis of PD can remain challenging, even at the motor stage. PET imaging can be used to confirm the clinical diagnosis. However, limitations in the robustness of predictive features extracted from the data and the costs associated with PET imaging restrict its application. Using blood metabolomics data as an additional information source may provide improved combined diagnostic models and/or an initial filter to decide on whether to apply PET imaging.
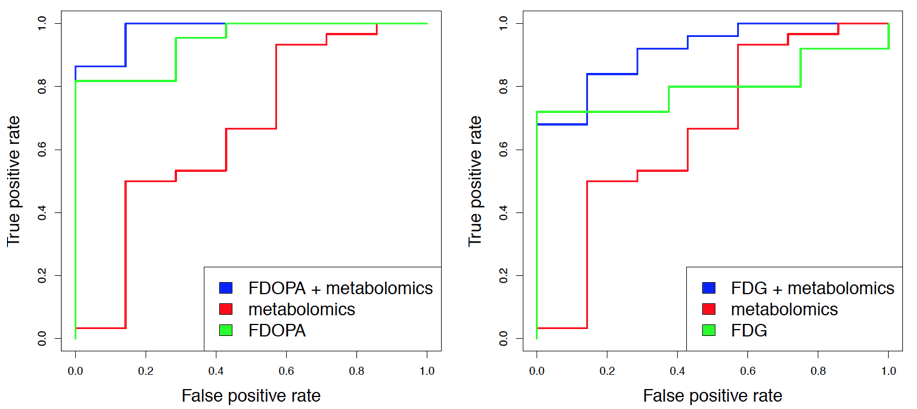
Methods: Metabolomics profiling of blood plasma samples using gas chromatography coupled to mass spectrometry (GCMS) was conducted in 60 IPD patients and 15 healthy controls. After pre-processing, these data were compared to neuroimaging data for subsets of the same individuals using FDOPA PET (44 patients and 14 controls) and FDG PET (51 patients and 15 controls). Machine learning models using linear support vector machines were trained on 50% of the data and evaluated on a 50% holdout test set using Receiver Operating Characteristic (ROC) curves. Next, standardized FDOPA and FDG PET intensity measurements were combined with those from the metabolomics data to build and evaluate sample classification models in the same manner as for the individual datasets.
Results: Both for the FDOPA and FDG PET data, the predictive performance given by the area under the ROC curve (AUC) was highest when combining imaging features with those from the metabolomics data (AUC for FDOPA + metabolomics: 0.98; AUC for FDG + metabolomics: 0.91). The performance was generally lower when using only the respective PET attributes (FDOPA: 0.94, FDG: 0.8) or only the metabolomics data (AUC: 0.66). [figure1]
Conclusions: GCMS metabolomics data from blood plasma samples contains predictive information for supervised classification of PD patients vs. controls which is independent FDOPA and FDG PET imaging data. Thus, blood metabolomics may add useful information to improve PD diagnosis via combined analyses with PET data. Alternatively, they may function as pre-screening filter when deciding on additional PET examinations in subjects at risk of developing PD. Further validation of the proposed machine learning models is required to assess the potential for generalization in various study cohorts.
To cite this abstract in AMA style:
E. Glaab, A. Greuel, J.P. Trezzi, C. Jäger, Z. Hodak, L. Timmermann, M. Tittgemeyer, N. Diederich. Combining PET imaging and blood metabolomics data to improve machine learning models for Parkinson’s disease diagnosis [abstract]. Mov Disord. 2018; 33 (suppl 2). https://www.mdsabstracts.org/abstract/combining-pet-imaging-and-blood-metabolomics-data-to-improve-machine-learning-models-for-parkinsons-disease-diagnosis/. Accessed December 19, 2025.« Back to 2018 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/combining-pet-imaging-and-blood-metabolomics-data-to-improve-machine-learning-models-for-parkinsons-disease-diagnosis/