Category: Parkinson's Disease: Genetics
Objective: To comprehensively investigate the genetic relationship between Parkinson’s disease (PD), its risk factors, and comorbidities by constructing a factor model of the shared genetic architecture.
Background: PD has multiple associated risk factors and comorbidities. Although genetic correlations have been identified for individual traits like smoking propensity in relation to PD, the overarching genetic underpinnings shared among PD risk factors remain underexplored.
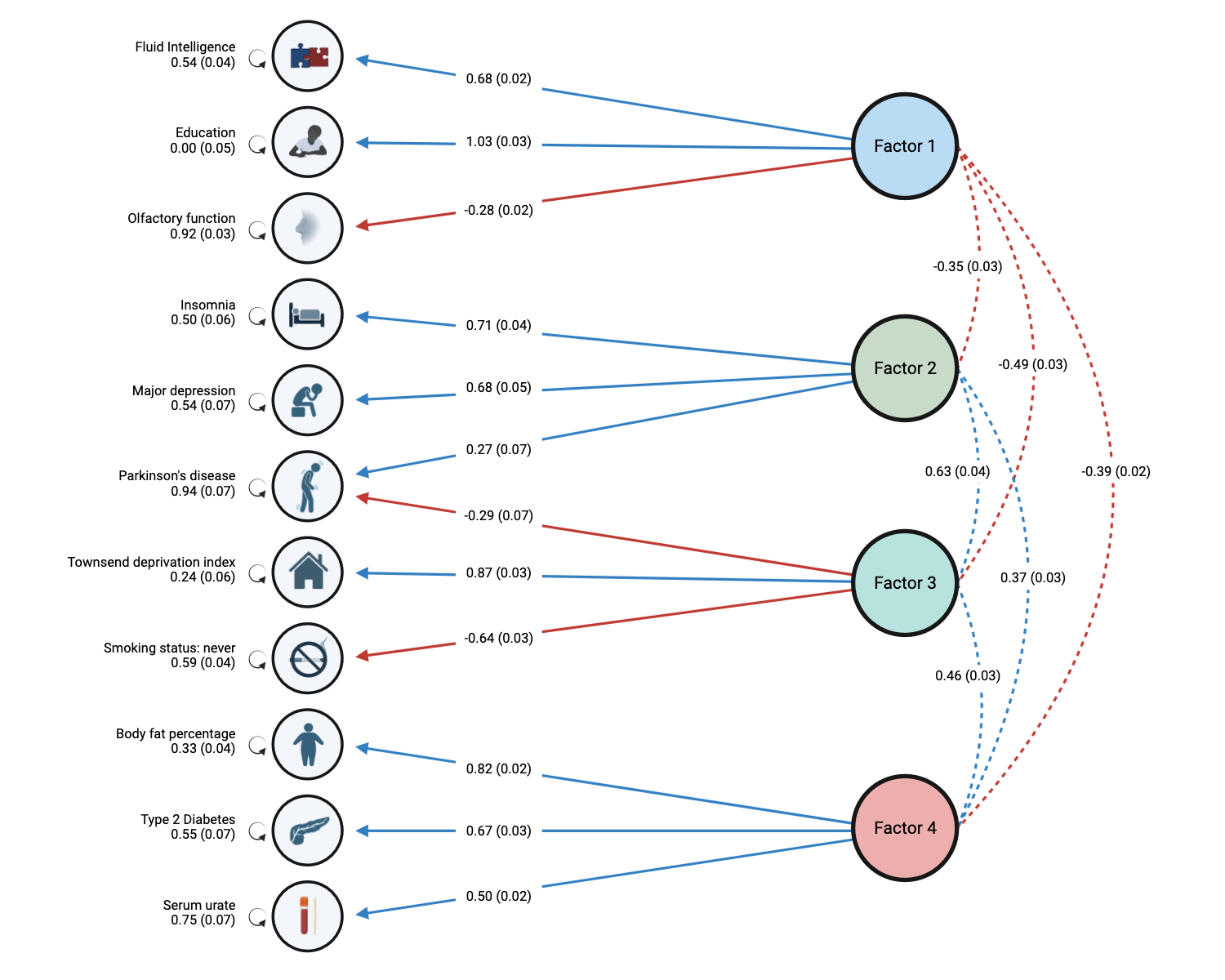
Method: Using Genomic Structural Equation Modeling (GSEM) [1], we examined the patterns of shared genetic liability across the PD and 10 related traits: education attainment, fluid intelligence, self-assessed ability to smell, insomnia, major depression, smoking status, deprivation status, body fat percentage, type-2 diabetes, and serum urate. Using factor analysis, we constructed a model of latent factors shared by these traits, then performed a multi-trait Genome-Wide Association Study (GWAS) to identify genetic variants associated with the latent factors. Stratified GSEM [2] was used to identify cell types and tissue-level enrichment associated with the factors, while transcriptome-wide SEM (T-SEM) [2] and Metascape gene set analysis [3] was used to link these factors with biological pathways. Finally, we validated the impact of the latent factors on PD risk using polygenic risk score (PRS) and mendelian randomization (MR) using an independent cohort of PD patients from Global Parkinson’s Genetics Program (GP2).
Results: Our model showed that PD and its related traits share genetic risk through 4 distinct but correlated latent factors. Stratified GSEM identified enrichment in brain tissues and specific neuronal cell types in these factors. Gene set analysis characterized the factors to have roles in cortical development, neuronal components, viral cellular stress responses, and metabolism. Multi-trait GWAS identified multiple novel loci and their impact on PD risk was validated using PRS and MR. Neuronal component factor (insomnia, major depression, PD) and metabolism factor (body fat percentage, type 2 diabetes, serum urate) showed the most significant associations.
Conclusion: Our model illustrates that many known risk factors and comorbidities of PD share common biological pathways that contribute to PD risk. These factors present mechanistic insights into common PD vulnerabilities and may be potential treatment targets for PD prevention.
Latent factor model of PD and 10 related traits
References: 1. Grotzinger AD, Rhemtulla M, de Vlaming R, Ritchie SJ, Mallard TT, Hill WD, et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav. 2019;3:513–25.
2. Grotzinger AD, de la Fuente J, Davies G, Nivard MG, Tucker-Drob EM. Transcriptome-wide and stratified genomic structural equation modeling identify neurobiological pathways shared across diverse cognitive traits. Nat Commun. 2022;13:6280.
3. Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523.
To cite this abstract in AMA style:
J. Kim, I. Foote, S. Bandres-Ciga, 23. Research Team, C. Marshall, A. Grotzinger3, A. Singleton, A. Brentnall, C. Blauwendraat, A. Noyce, G. Genetics Program. Latent Genetic Architecture of Parkinson’s Disease Risk factors and Comorbidities [abstract]. Mov Disord. 2024; 39 (suppl 1). https://www.mdsabstracts.org/abstract/latent-genetic-architecture-of-parkinsons-disease-risk-factors-and-comorbidities/. Accessed December 25, 2025.« Back to 2024 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/latent-genetic-architecture-of-parkinsons-disease-risk-factors-and-comorbidities/