Objective: We aimed to identify potentially pathogenic genomic variations associated with dystonia in Asian Indian patients using whole exome sequencing.
Background: Genomic variations associated with dystonia in the Asian Indian population remain largely unknown as they are underrepresented in the larger genetic studies.
Method: The Indian Movement Disorder Registry and Biobank is a multi-centric clinical registry and DNA biorepository for Indian patients with suspected genetic movement disorders. As part of this registry, our center enrolled 570 probands and affected/unaffected family members with dystonia from Sep 2018- Sep 2021. Clinical and demographic data were captured and stored on a REDCap platform. All patients underwent standardized videotaped neurological examinations. We performed whole exome sequencing (WES) on DNA specimens obtained from 254 probands with isolated, combined or complex dystonia. DNA libraries were sequenced to depths of 80-100X on an Illumina platform. We followed the GATK best practices framework for variant calling and prioritized variants according to prespecified criteria. Finally, variants were classified according to ACMG guidelines.
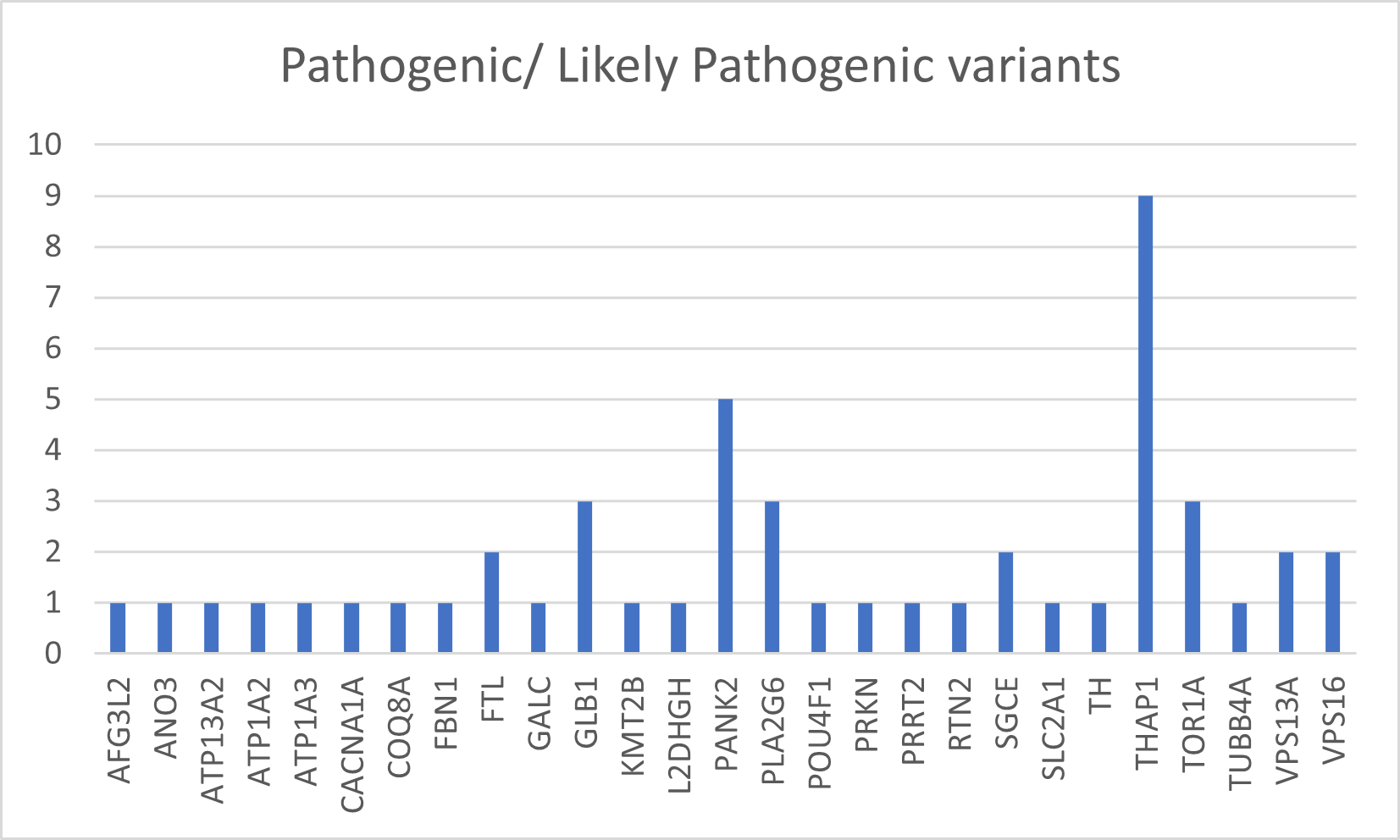
Results: The mean age of the WES cohort was 35.1±16.2 years and age at onset was 27.2±17.4 years. Dystonia was early onset in 43.3% and generalized in 23.2%. Family history was positive for dystonia or another movement disorder in 35.8%. 68.5% had isolated dystonia, 11.8% were combined and 18.9% were complex phenotypes. WES identified pathogenic/ likely pathogenic variants in 49 patients (19.2%) including 9 novel variants in known dystonia genes. Mutations in THAP1 were most common followed by SCGE, VPS16, PANK2, GLB1, FTL, TOR1A, TUBB4A, ATP13A2, ANO3, TH, FBN1, RTN2, COQ8A and other genes (Figure 1). Variants of uncertain significance were identified in additional 106 participants- most commonly in the COL6A3 gene.
Conclusion: WES identified potentially pathogenic variations in 19.2% patients in this cohort. Several genes known to be associated with dystonia were identified in this cohort of Indian dystonia, with novel variants in some. THAP1 was the most frequently encountered dystonia associated gene. This is currently the largest cohort of genetically defined dystonia from the Asian Indian population; such ethnicity specific genetic information from underrepresented populations may help to address the missing heritability in monogenic disorders.
To cite this abstract in AMA style:
R. Rajan, A. Saini, R. Mewara, B. Verma, D. Radhakrishnan, A. Agarwal, E. A, A. Gupta, V. V Y, M. Singh, R. Bhatia, I. Singh, R. Mir, F. Mohammad, B. B K, V. Scaria, A. Srivastava, M V. Srivastava. Genetic spectrum of monogenic dystonia in Asian Indian patients [abstract]. Mov Disord. 2023; 38 (suppl 1). https://www.mdsabstracts.org/abstract/genetic-spectrum-of-monogenic-dystonia-in-asian-indian-patients/. Accessed December 28, 2025.« Back to 2023 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/genetic-spectrum-of-monogenic-dystonia-in-asian-indian-patients/