Category: Ataxia
Objective: To investigate variations in regulatory regions of the FXN gene hampering its expression in Friedreich’s Ataxia (FRDA) patients. To investigate variations in regulatory regions of the FXN gene hampering its expression in Friedreich’s Ataxia (FRDA) patients.
Background: Highly conserved regulatory regions acting as enhancers/repressors are located upstream of the FXN gene.1 These regions have a direct role in regulating FXN expression. Any variation or mutation can alter the binding of transcription factors affecting gene expression.
Method: Homozygous FRDA patients were enrolled in ‘Ataxia Clinic’ from the Department of Neurology, AIIMS, New Delhi. Healthy controls (genetically normal for GAA repeats) were also enrolled. Regulatory regions of interest (Region A- Chr9:69030494-69030759; Region B-chr9:9026436-69026607; Region C- chr9:69023464-69023914) were amplified using standard PCR and purified. The variations were identified using high throughput sequencing on a 2*150bp paired-end run using the NovaSeq 6000 instrument. Data analysis was carried out on the GRCh38.p13 database using Genome Analysis Toolkit (GATK) v4.2.4.0. Results were verified using Sanger sequencing. The eQTL (expression Quantitative Trait Loci) analysis using GTEx.
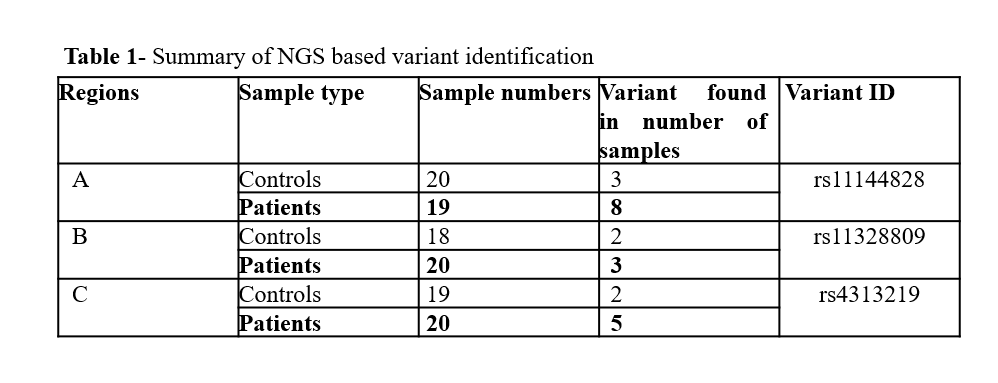
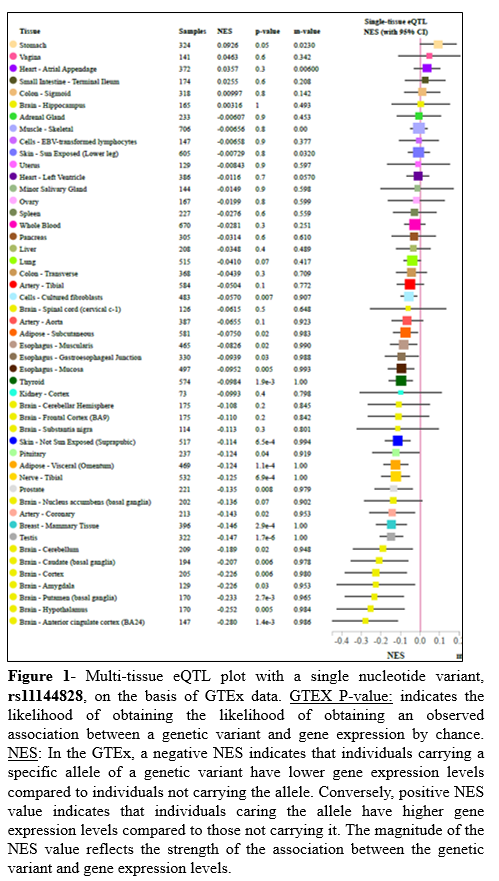
Results: Three regulatory regions of 20 healthy controls and 19 FRDA patients were investigated for variations. The variation G>A/C (rs11144828) was found in region-A, del(A)5 indel (rs11328809) in region-B and variation T>A (rs4313219) was found in region C (Table-1). None of these variations was reported in ClinVar database. The GTEx analysis showed that the variant rs11144828 affected frataxin expression in several tissues, especially in different regions of the brain (p<0.05) with a high correlation score (m-value ~1.0). Negative NES values in Figure-1 indicates that patients carrying the variant have lower gene expression levels compared to individuals not carrying the allele. This indicates that the variant identified in the present study can affect the frataxin levels of FRDA patients (containing low GAA repeats, as a number of GAA repeats is directly linked to severity and early onset).
Conclusion: Variants are present in regulatory regions of FXN gene. The presence of rs11144828 in region-A demonstrates its propensity to deregulate FXN levels in patients. Further analysis is required to investigate if any transcription factor binds it.
References: Puspasari et al. Long range regulation of human FXN gene expression. PLoS One. 2011;6(7):e22001.
To cite this abstract in AMA style:
V. Swarup, H. Singh, V. Kumar, D. Gupta, A. Ahuja, I. Singh, R. Rajan, A. Srivastava. Targeted sequencing of regulatory regions shows potential SNVs affecting FXN gene expression in Friedreich’s ataxia patients [abstract]. Mov Disord. 2023; 38 (suppl 1). https://www.mdsabstracts.org/abstract/targeted-sequencing-of-regulatory-regions-shows-potential-snvs-affecting-fxn-gene-expression-in-friedreichs-ataxia-patients/. Accessed December 23, 2025.« Back to 2023 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/targeted-sequencing-of-regulatory-regions-shows-potential-snvs-affecting-fxn-gene-expression-in-friedreichs-ataxia-patients/