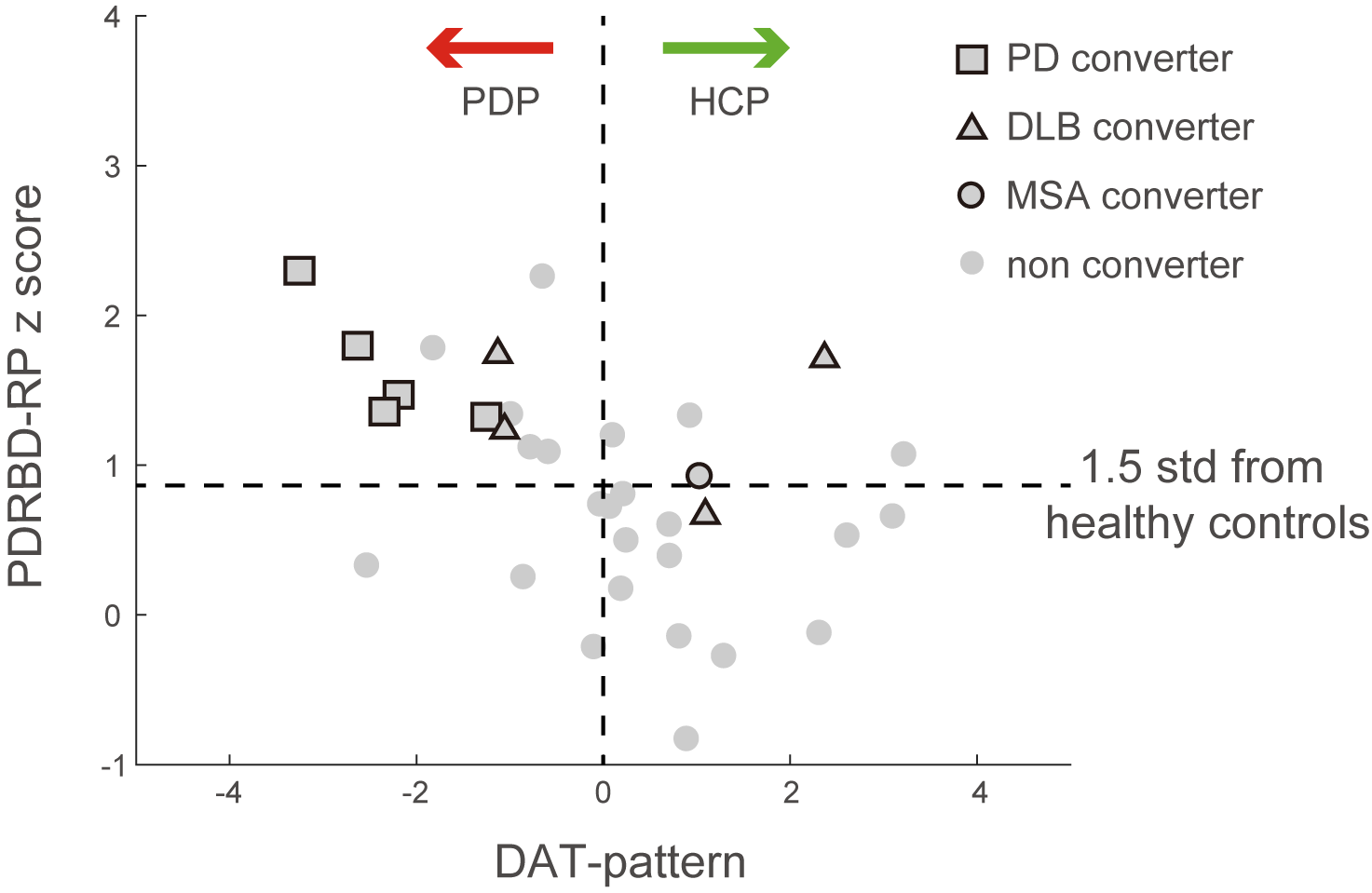
Objective: To elucidate the role of Parkinson’s disease (PD)-related brain metabolic patterns as a biomarker in isolated rapid-eye-movement sleep behavior disorder (iRBD) for clinical correlation and future disease conversion.
Background: Disease-related metabolic patterns derived from Parkinson’s disease (PD-RP) and de-novo PD with probable RBD (dnPDRBD-RP) have been proposed as a biomarker in iRBD. However, the direct comparisons between the metabolic patterns derived from the long-standing and de novo PD patients (PD-RP vs. dnPDRBD-RP) in terms of clinical biomarker correlation and predictability of future phenoconversion have not been investigated.
Method: We prospectively enrolled 30 iRBD patients, 25 de novo PD patients with a premorbid history of RBD, 21 long-standing PD patients on stable treatment and 24 healthy controls. Participants underwent 18F-Fluorodeoxyglucose (FDG) PET scans and clinical tests for olfaction and cognition, and the Movement disorders society-Unified PD Rating Scale (MDS-UPDRS) evaluation. From FDG-PET scans, we derived two metabolic patterns from the long-standing PD group (PD-RP) and PD de novo group (dnPDRBD-RP). Subsequently, we calculated the PD-RP and dnPDRBD-RP expression scores in iRBD patients. We validated the two metabolic patterns in each PD group and our separate iRBD cohort (n=14).
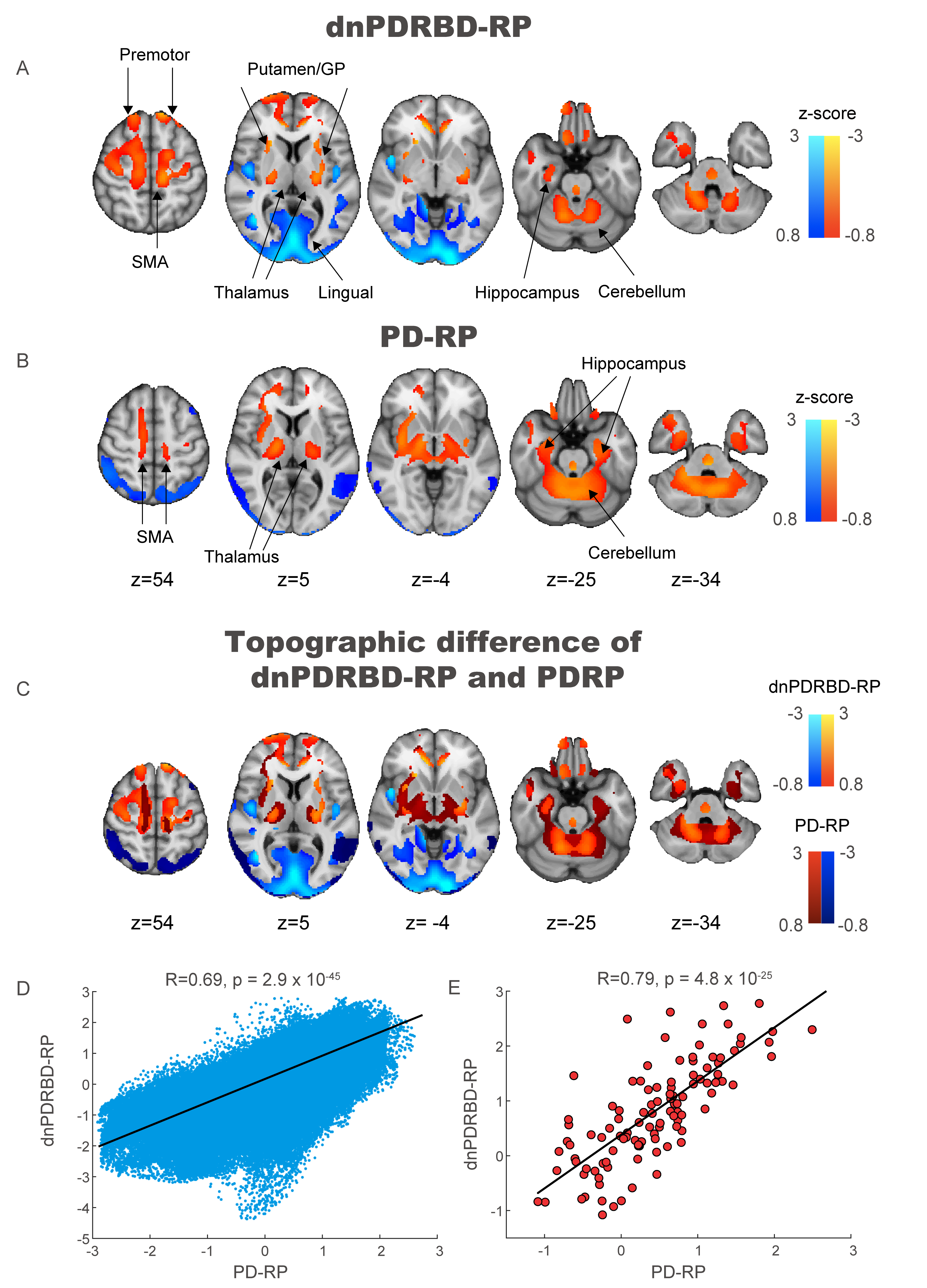
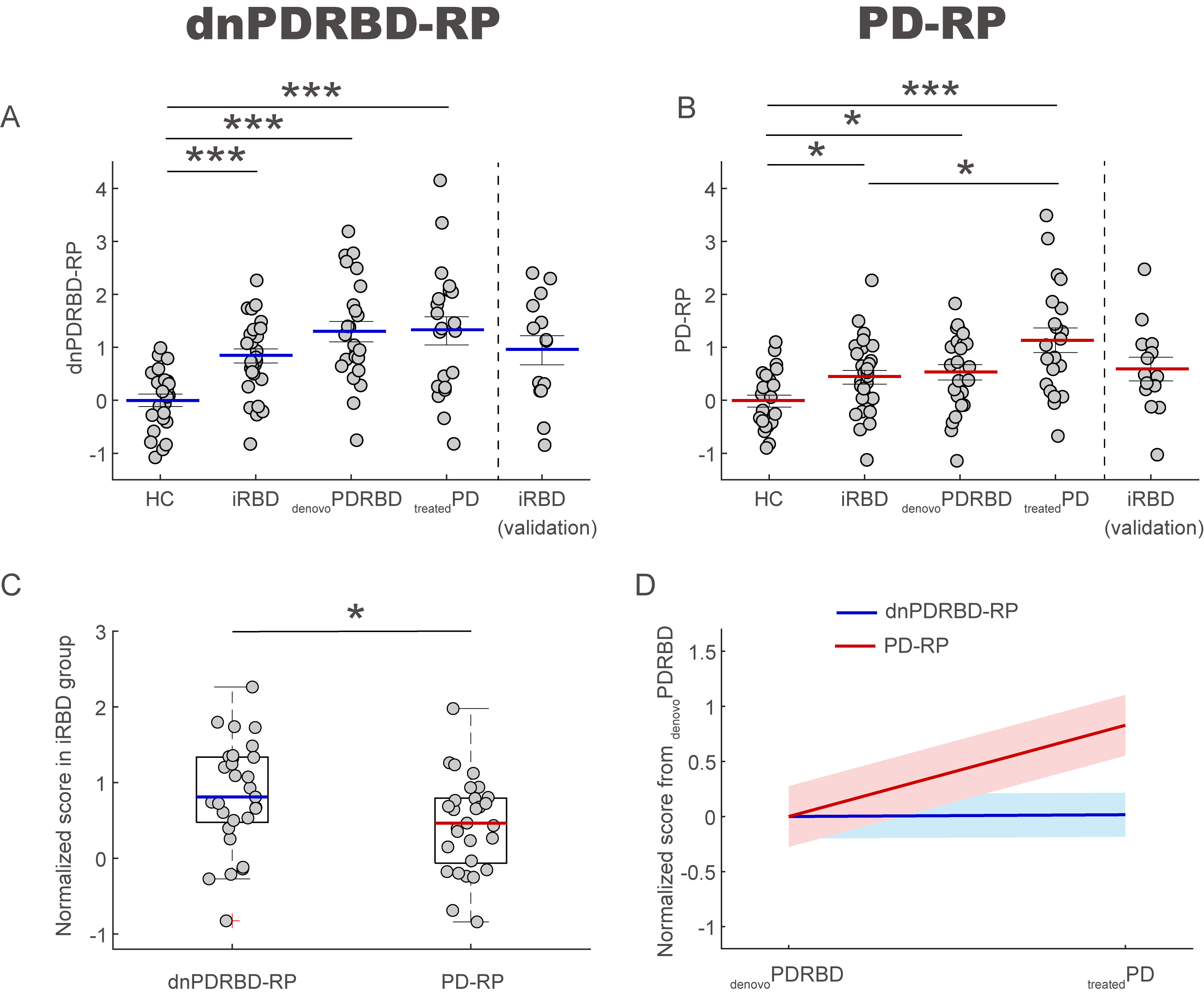
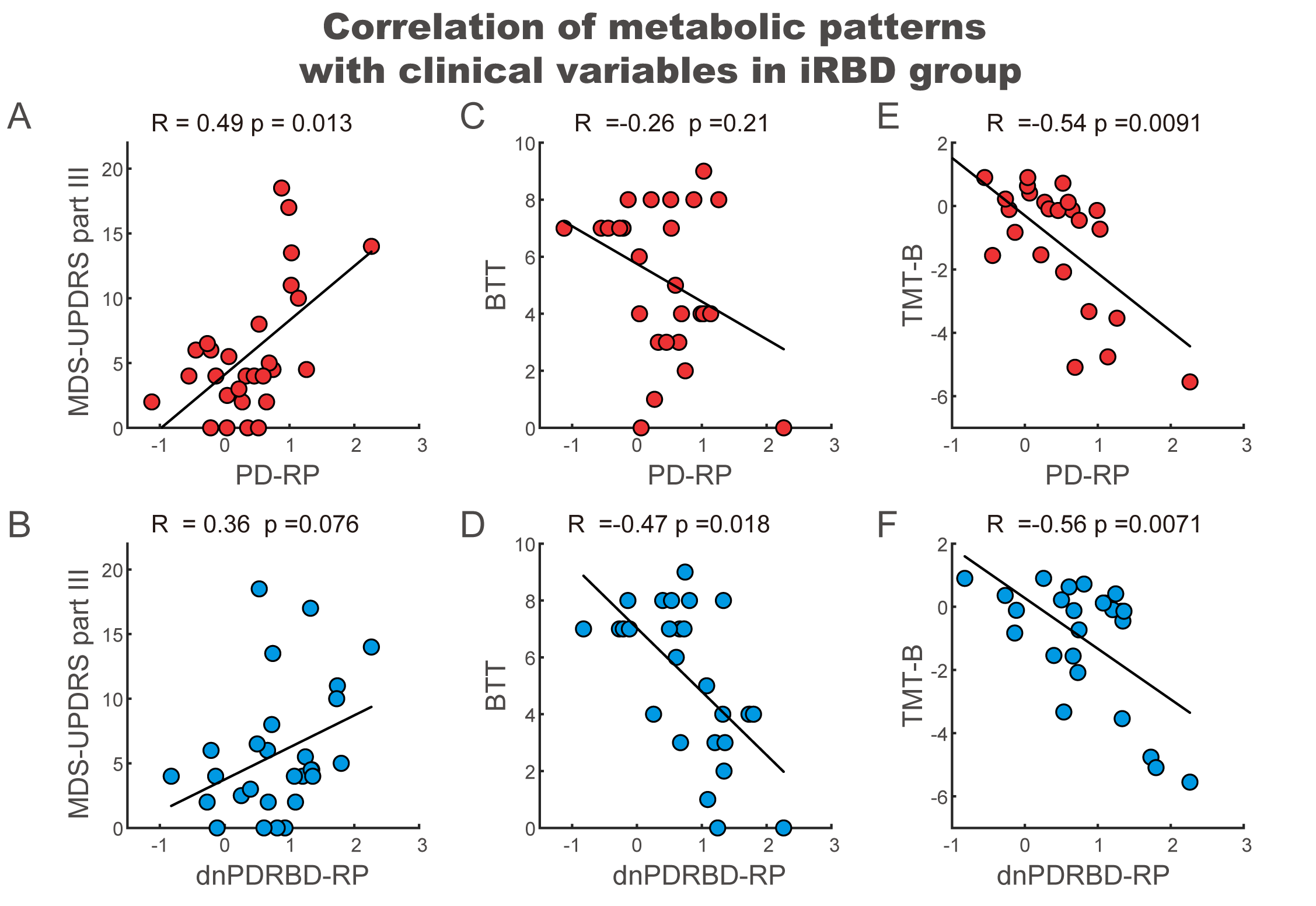
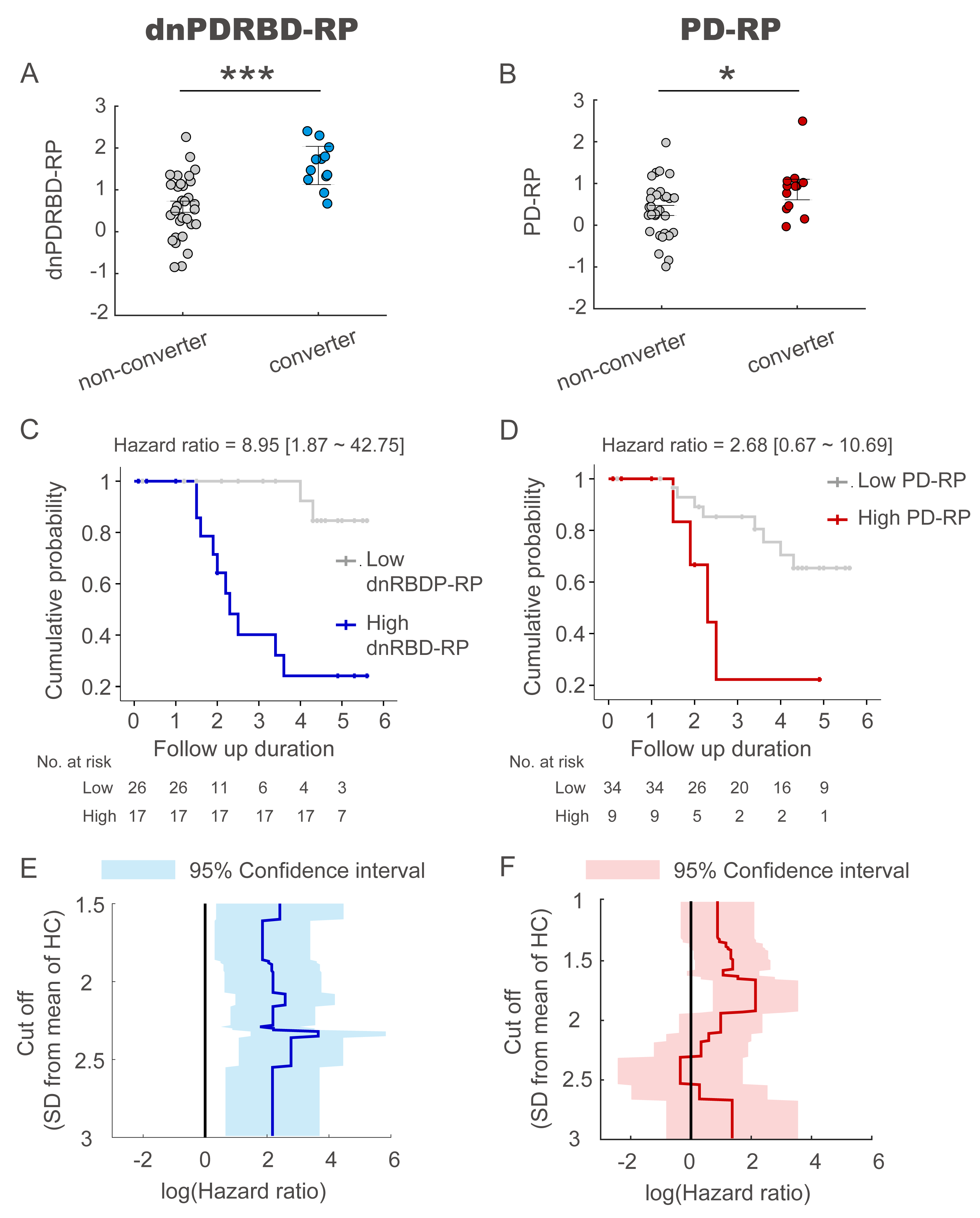
Results: The two patterns significantly correlated with each other and were spatially overlapping yet distinct. The MDS-UPDRS motor scores significantly correlated with PD-RP (p = 0.013) but insignificant with dnPDRBD-RP scores (p = 0.076). In contrast, dnPDRBD-RP correlated with olfaction in butanol threshold test (p = 0.018) in iRBD subjects, but PD-RP did not (p = 0.21). High dnPDRBD-RP expression in iRBD patients predicted future phenoconversion with all cut-off ranges from 1.5 to 3 standard deviations of the control value, whereas predictability of PD-RP was only significant in a partial range of cut-off.
Conclusion: The present study is the first comparative analysis on two PD-related metabolic patterns of dnPDRBD-RP and PD-RP in a prospective iRBD cohort. dnPDRBD-RP shares the metabolic pattern of PD-RP, yet distinct. Overall, dnPDRBD-RP better reflects prodromal clinical features of PD and better predicts future phenoconversion in iRBD as it was derived from de-novo PD with a history of preceding RBD which may incorporate the compensatory change of the prodromal stage of PD.
References: 1. Heinzel S, Berg D, Gasser T, et al. Update of the MDS research criteria for prodromal Parkinson’s disease. Mov Disord 2019;34:1464-1470. 2. Postuma RB, Iranzo A, Hu M, et al. Risk and predictors of dementia and parkinsonism in idiopathic REM sleep behaviour disorder: a multicentre study. Brain 2019;142:744-759. 3. Iranzo A, Fernandez-Arcos A, Tolosa E, et al. Neurodegenerative disorder risk in idiopathic REM sleep behavior disorder: study in 174 patients. PLoS One 2014;9:e89741. 4. Shin JH, Lee J-Y, Kim Y-K, et al. Longitudinal change in dopamine transporter availability in idiopathic REM sleep behavior disorder. Neurology 2020;Online ahead of print; doi: 10.1212/WNL.0000000000010942. 5. Ma Y, Tang C, Spetsieris PG, Dhawan V, Eidelberg D. Abnormal metabolic network activity in Parkinson’s disease: test-retest reproducibility. J Cereb Blood Flow Metab 2007;27:597-605. 6. Holtbernd F, Ma Y, Peng S, et al. Dopaminergic correlates of metabolic network activity in Parkinson’s disease. Hum Brain Mapp 2015;36:3575-3585. 7. Huang C, Tang C, Feigin A, et al. Changes in network activity with the progression of Parkinson’s disease. Brain 2007;130:1834-1846. 8. Ko JH, Lee CS, Eidelberg D. Metabolic network expression in parkinsonism: Clinical and dopaminergic correlations. J Cereb Blood Flow Metab 2017;37:683-693. 9. Schindlbeck KA, Eidelberg D. Network imaging biomarkers: insights and clinical applications in Parkinson’s disease. Lancet Neurol 2018;17:629-640. 10. Wu P, Wang J, Peng S, et al. Metabolic brain network in the Chinese patients with Parkinson’s disease based on 18F-FDG PET imaging. Parkinsonism Relat Disord 2013;19:622-627. 11. Tomse P, Jensterle L, Grmek M, et al. Abnormal metabolic brain network associated with Parkinson’s disease: replication on a new European sample. Neuroradiology 2017;59:507-515. 12. Meles SK, Renken RJ, Pagani M, et al. Abnormal pattern of brain glucose metabolism in Parkinson’s disease: replication in three European cohorts. Eur J Nucl Med Mol Imaging 2020;47:437-450. 13. Kogan RV, Janzen A, Meles SK, et al. Four-Year Follow-up of [(18) F]Fluorodeoxyglucose Positron Emission Tomography-Based Parkinson’s Disease-Related Pattern Expression in 20 Patients With Isolated Rapid Eye Movement Sleep Behavior Disorder Shows Prodromal Progression. Mov Disord 2020. 14. Yoon EJ, Lee JY, Nam H, et al. A New Metabolic Network Correlated with Olfactory and Executive Dysfunctions in Idiopathic Rapid Eye Movement Sleep Behavior Disorder. J Clin Neurol 2019;15:175-183. 15. Holtbernd F, Gagnon JF, Postuma RB, et al. Abnormal metabolic network activity in REM sleep behavior disorder. Neurology 2014;82:620-627. 16. Sixel-Doring F, Trautmann E, Mollenhauer B, Trenkwalder C. Associated factors for REM sleep behavior disorder in Parkinson disease. Neurology 2011;77:1048-1054. 17. Olson EJ, Boeve BF, Silber MH. Rapid eye movement sleep behaviour disorder: demographic, clinical and laboratory findings in 93 cases. Brain 2000;123 ( Pt 2):331-339. 18. Postuma RB, Gagnon JF, Vendette M, Charland K, Montplaisir J. REM sleep behaviour disorder in Parkinson’s disease is associated with specific motor features. J Neurol Neurosurg Psychiatry 2008;79:1117-1121. 19. Vendette M, Gagnon JF, Décary A, et al. REM sleep behavior disorder predicts cognitive impairment in Parkinson disease without dementia. Neurology 2007;69:1843-1849. 20. Lim JS, Shin SA, Lee JY, Nam H, Lee JY, Kim YK. Neural substrates of rapid eye movement sleep behavior disorder in Parkinson’s disease. Parkinsonism Relat Disord 2016;23:31-36. 21. Wu P, Yu H, Peng S, et al. Consistent abnormalities in metabolic network activity in idiopathic rapid eye movement sleep behaviour disorder. Brain 2014;137:3122-3128. 22. Huber M, Beyer L, Prix C, et al. Metabolic Correlates of Dopaminergic Loss in Dementia with Lewy Bodies. Mov Disord 2020;35:595-605. 23. Lee J-Y, Yoon EJ, Kim YK, et al. Nonmotor and Dopamine Transporter Change in REM Sleep Behavior Disorder by Olfactory Impairment. J Mov Disord 2019;12:103-112. 24. Schindlbeck KA, Lucas-Jimenez O, Tang CC, et al. Metabolic Network Abnormalities in Drug-Naive Parkinson’s Disease. Mov Disord 2020;35:587-594.
To cite this abstract in AMA style:
JH. Shin, JY. Lee, YK. Kim, HJ. Kim, EJ. Yoon, HW. Nam, B. Jeon. Metabolic patterns and neurodegeneration in isolated REM sleep behavior disorder [abstract]. Mov Disord. 2021; 36 (suppl 1). https://www.mdsabstracts.org/abstract/metabolic-patterns-and-neurodegeneration-in-isolated-rem-sleep-behavior-disorder/. Accessed April 2, 2025.« Back to MDS Virtual Congress 2021
MDS Abstracts - https://www.mdsabstracts.org/abstract/metabolic-patterns-and-neurodegeneration-in-isolated-rem-sleep-behavior-disorder/