Category: Parkinson's Disease: Genetics
Objective: To identify genetic modifiers and interactors that may modulate known risk variants in the SNCA, TMEM175 and MAPT loci in Parkinson’s disease (PD).
Background: The most recent PD genome-wide association study (GWAS) identified 90 independent genetic associations in 78 loci associated with PD risk [1]. Possible genetic modifiers of risk and onset of GBA-associated PD have been identified [2], however interaction between the remaining PD genetic risk loci remains unknown.
Method: To perform stratified GWASs and identify potential modifiers of some of the strongest genetic risk factors in PD, we used the International Parkinson’s Disease Genomics Consortium genotyping data, including a total of 32,338 participants (PD=14,671, controls=17,667). We stratified our cohort for carriers and non-carriers of the PD GWAS hits SNCA rs356182, TMEM175 rs34311866 and the MAPT H2 haplotype tagging variant rs8070723. Six case-control GWASs were performed in carriers and non-carriers of the selected SNPs using adjusted logistic regression models.
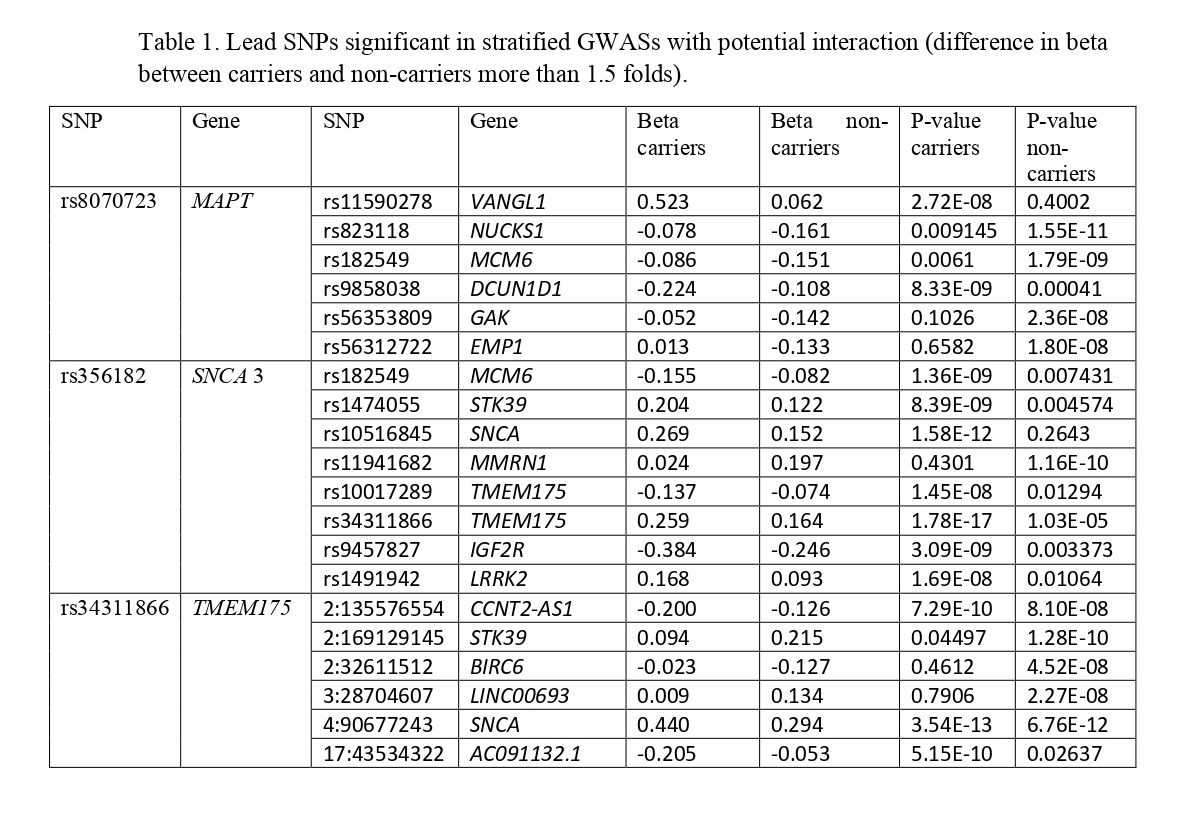
Results: We identified six independent risk loci for MAPT H2 haplotype carriers and ten for non-carriers surpassing GWAS significance threshold (p=5×10−8) [figure 1]. The SNP rs11590278 near VANGL1 demonstrated interaction with MAPT H2 haplotype (OR= 1.38, p= 4.3E-05). MAPT H1/H1 haplotype had potential interaction with rs56312722 near EMP1 [table 1]. Four more GWA significant lead SNPs were found significant, with at least 1.5-fold difference in effect size (beta) between the haplotypes [table 1].
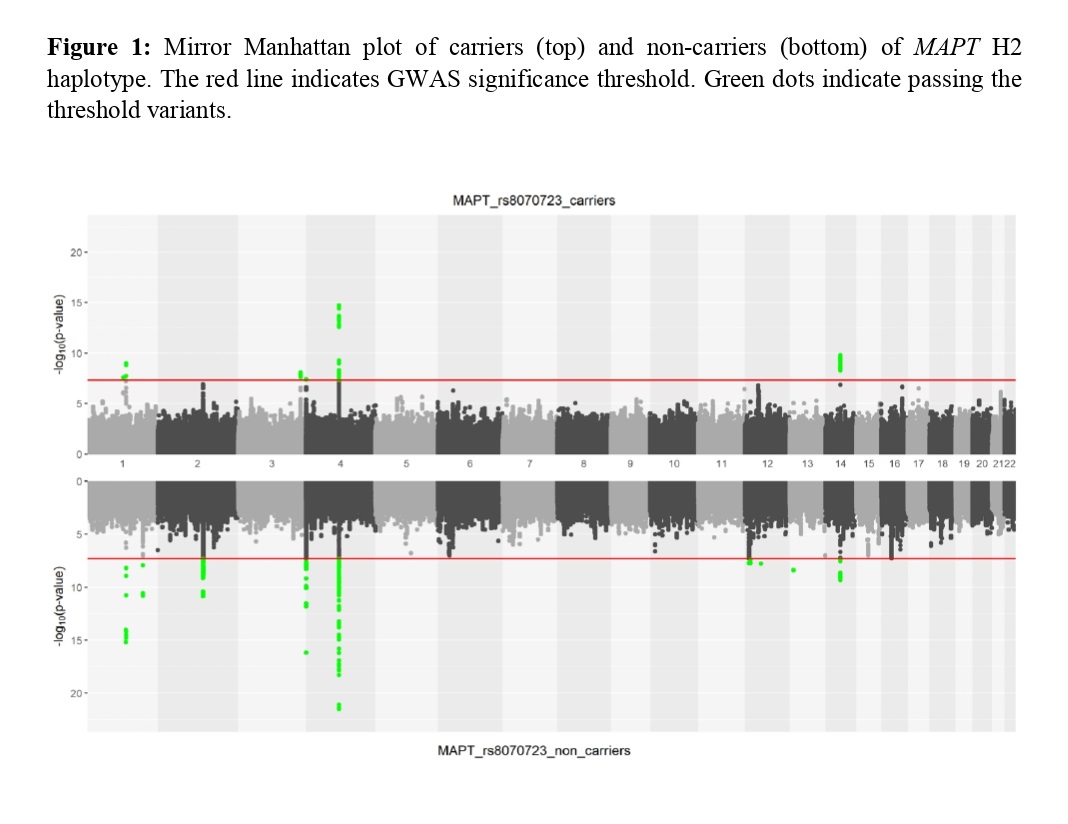
We found 11 genomic risk loci for SNCA 3’ rs356182 carriers and three for non-carriers [figure 2]. We identified seven significant lead SNPs in SNCA rs356182 carriers which had at least 1.5-fold difference in effect size with non-carriers. Interestingly, we found that rs11941682 in MMRN1 is associated with PD in non-carriers of rs356182 and also in LD with rs356182 (D’= 0.84) [table 1].
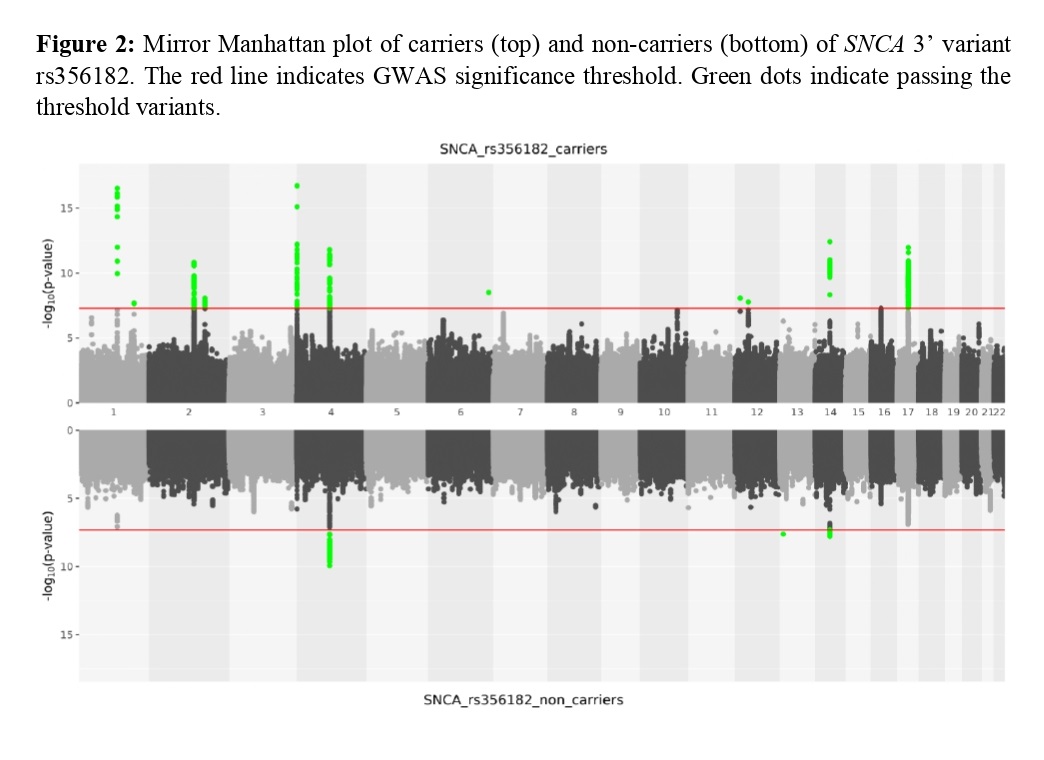
In the TMEM175 stratified GWAS, five genomic risk loci were found for rs4952211 carriers and 12 for non-carriers [figure 3]. Carriers of rs4952211 had two and non-carriers had three significant lead SNPs with meaningful differences in effect size [table 1].
Conclusion: Our results suggest possible interactions of different SNPs with PD GWAS top hit loci. Functional studies are required to determine if and how these potential interactions are important in the mechanisms underlying PD.
References: [1] Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, Tan M, Kia DA, Noyce AJ, Xue A (2019) Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. The Lancet Neurology 18, 1091-1102. [2] Blauwendraat C, Reed X, Krohn L, Heilbron K, Bandres-Ciga S, Tan M, Gibbs JR, Hernandez DG, Kumaran R, Langston R (2020) Genetic modifiers of risk and age at onset in GBA associated Parkinson’s disease and Lewy body dementia. Brain 143, 234-248.
To cite this abstract in AMA style:
K. Senkevich, S. Bandres-Ciga, E. Yu, L. Krohn, Z. Gan-Or. Stratified genome-wide association study revealed novel interactions for the top hit Parkinson’s disease loci. [abstract]. Mov Disord. 2021; 36 (suppl 1). https://www.mdsabstracts.org/abstract/stratified-genome-wide-association-study-revealed-novel-interactions-for-the-top-hit-parkinsons-disease-loci/. Accessed December 29, 2025.« Back to MDS Virtual Congress 2021
MDS Abstracts - https://www.mdsabstracts.org/abstract/stratified-genome-wide-association-study-revealed-novel-interactions-for-the-top-hit-parkinsons-disease-loci/