Category: Huntington's Disease
Objective: This study combines methylation data from the two largest EWAS studies in Huntington’s Disease (HD) human brain tissue [1,2]. Combined, this study yields the most robust EWAS to date. This analysis is furthered by using libraries of cell-specific methylation data to generate the first estimation of cell-type proportions in HD brains.
Background: HD is caused by a mutation in the HTT (huntingtin) gene on chromosome 4 leading to CAG repeat and polyglutamine expansion in the huntingtin protein. HD has shown to have aberrant mRNA expression, transcriptional dysregulation in HD cell lines, mice and human brain tissue [3.4.5]. Previous epigenome wide association studies (EWAS) have shown epigenetic aging in peripheral blood and HD brain tissue samples. Studies using brain tissue have differed in their EWAS findings. Separately, HD has been characterized as degeneration of GABAergic neurons in the caudate nucleus and in the cortex.
Method: Publicly available DNA methylation data from two studies using cortical, basal ganglia, and cerebellum samples from human HD patient autopsies were arrayed via the Illumina450k platform were combined and re-analyzed. We generated a CNS DNA methylation library using information from isolated cells using surface or nuclear specific markers. Differential CpG sites were then identified using limma for each cell type and then used to deconvolve whole brain tissue samples into cell-type proportion estimations.
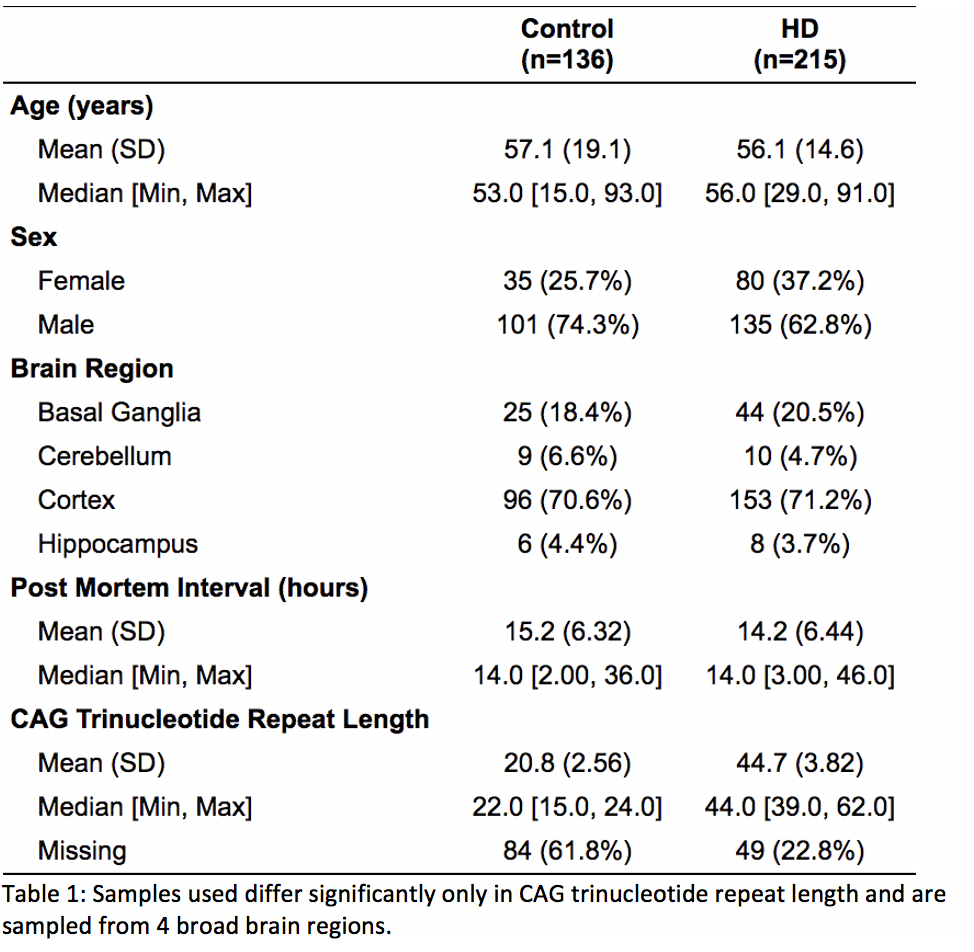
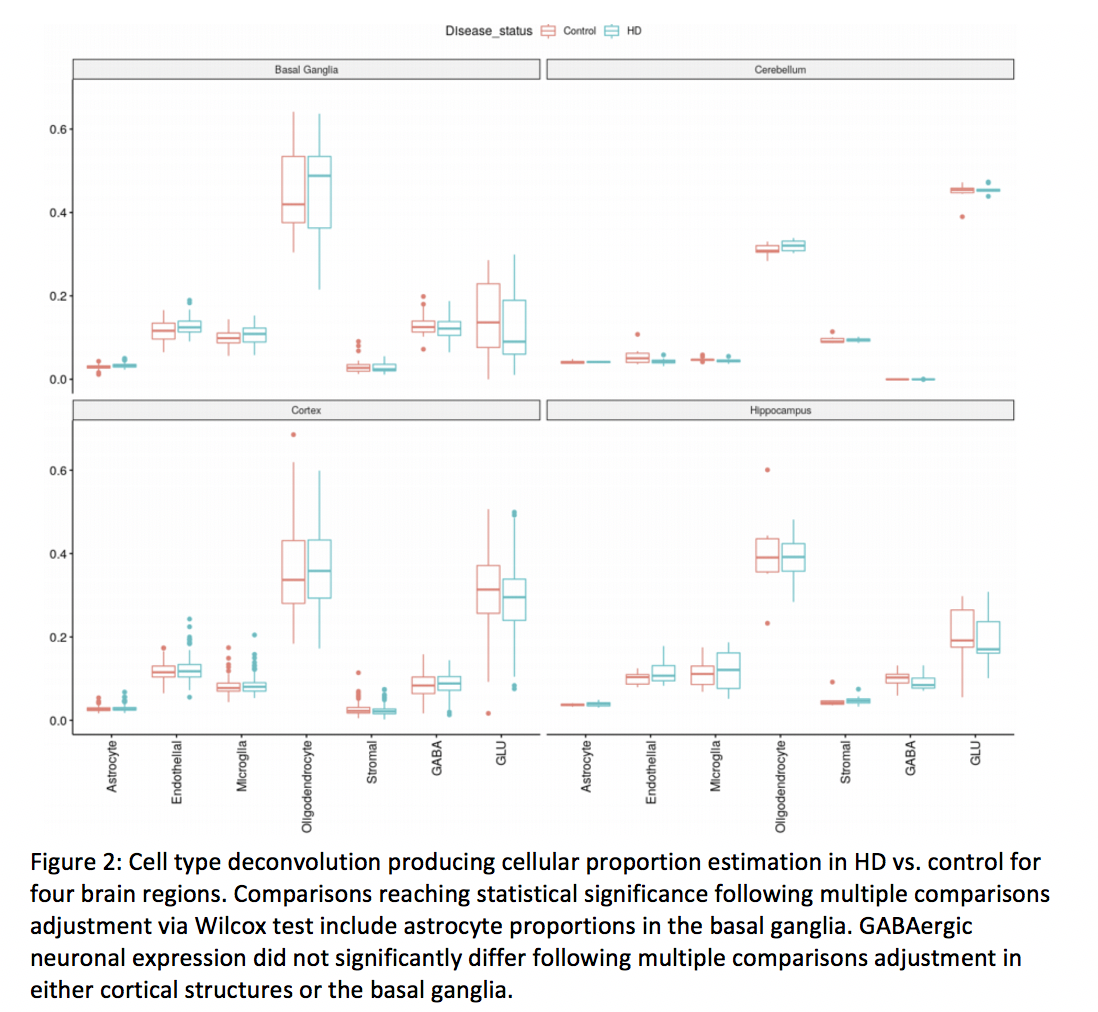
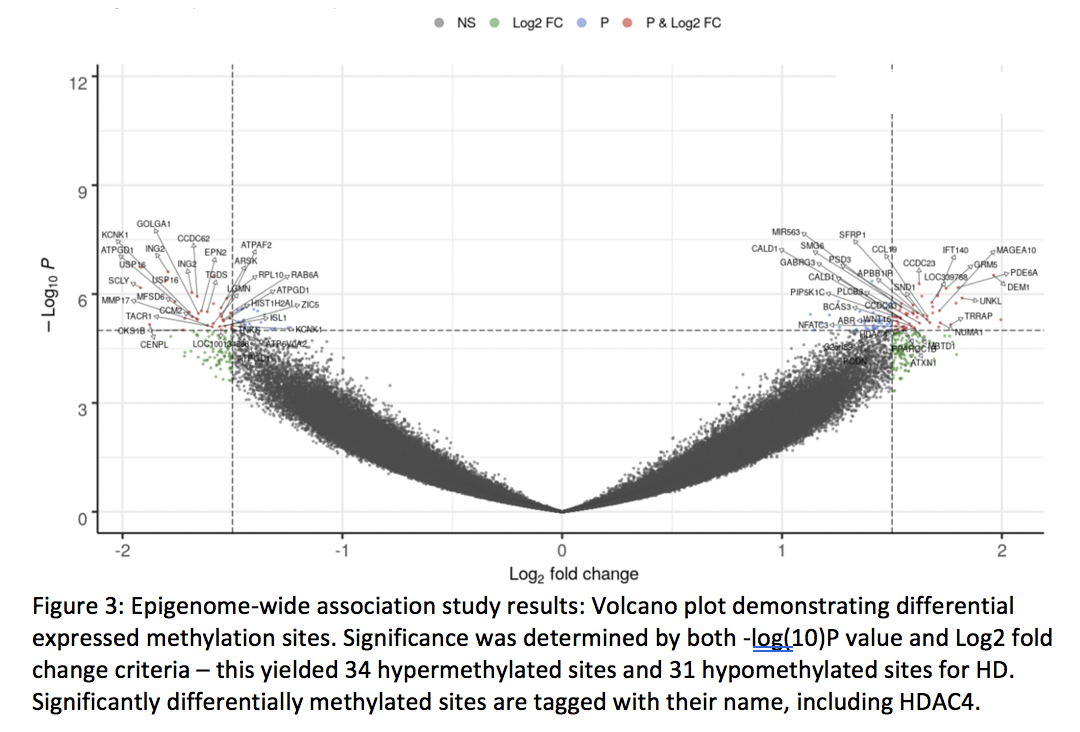
Results: This yielded 351 brain tissue samples (215=HD) from 60 subjects (33 = HD). Samples did not differ significantly across age, sex, post-mortem interval or brain regions sampled; however did differ significantly in CAG trinucleotide length. EWAS revealed 65 differentially methylated sites. Among those sites, HDAC4 was differentially methylated. Deconvolution of neural tissue methylation data did not reveal significant loss of GABAergic neurons in the basal ganglia of HD samples, nor was CAG repeat length associated with relative loss of neurons in HD samples.
Conclusion: This study supports that there are DNA methylation alterations in HD. Further characterization of how the polyglutamine expansion could affect specific areas of the GABAergic system in the CNS will require larger sample sizes. Similarly, experimental HDAC inhibitors could benefit from additional complementary DNA methylation epigenetic modifying HD therapies.
References: 1: Horvath S, Langfelder P, Kwak S, Aaronson J, Rosinski J, Vogt TF, Eszes M, Faull RL, Curtis MA, Waldvogel HJ, Choi OW, Tung S, Vinters HV, Coppola G, Yang XW. Huntington’s disease accelerates epigenetic aging of human brain and disrupts DNA methylation levels. Aging (Albany NY). 2016 Jul;8(7):1485-512. doi:10.18632/aging.101005. PubMed PMID: 27479945; PubMed CentralPMCID:PMC4993344. 2: De Souza RA, Islam SA, McEwen LM, Mathelier A, Hill A, Mah SM, Wasserman WW, Kobor MS, Leavitt BR. DNA methylation profiling in human Huntington’s disease brain. Hum Mol Genet. 2016 May 15;25(10):2013-2030. Epub 2016 Mar 6. PubMed PMID:26953320. 3: Cha JH. Transcriptional dysregulation in Huntington’s disease. Trends Neurosci. 2000 Sep;23(9):387-92. Review. PubMed PMID: 10941183. 4: Hodges A, Strand AD, Aragaki AK, Kuhn A, Sengstag T, Hughes G, Elliston LA,Hartog C, Goldstein DR, Thu D, Hollingsworth ZR, Collin F, Synek B, Holmans PA, Young AB, Wexler NS, Delorenzi M, Kooperberg C, Augood SJ, Faull RL, Olson JMJones L, Luthi-Carter R. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum Mol Genet. 2006 Mar 15;15(6):965-77. Epub 2006 Feb 8. PubMed PMID: 16467349 5: HD iPSC Consortium. Developmental alterations in Huntington’s disease neural cells and pharmacological rescue in cells and mice. Nat Neurosci. 2017 May;20(5):648-660. doi: 10.1038/nn.4532. Epub 2017 Mar 20. PubMed PMID: 28319609; PubMed Central PMCID: PMC5610046.
To cite this abstract in AMA style:
A. Glaser, J. Levy, Z. Zhang, L. Salas. Using Human Neural Tissue Methylation to Decipher Epigenetic Characteristics and Cell Type Pathologies in Huntington’s Disease [abstract]. Mov Disord. 2021; 36 (suppl 1). https://www.mdsabstracts.org/abstract/using-human-neural-tissue-methylation-to-decipher-epigenetic-characteristics-and-cell-type-pathologies-in-huntingtons-disease/. Accessed February 6, 2026.« Back to MDS Virtual Congress 2021
MDS Abstracts - https://www.mdsabstracts.org/abstract/using-human-neural-tissue-methylation-to-decipher-epigenetic-characteristics-and-cell-type-pathologies-in-huntingtons-disease/