Session Information
Date: Wednesday, September 25, 2019
Session Title: Neuroimaging
Session Time: 1:15pm-2:45pm
Location: Les Muses Terrace, Level 3
Objective: This study investigates differences in brain network organization in patients with Parkinson’s disease (PD) with genetic risk factors relative to sporadic PD patients and healthy control subjects (HC).
Background: PD patients with genetic risk factors have been clinically indistinguishable from sporadic cases, although the two most common genetic risk factors (LRRK2 G2019S and GBA) seem to produce a more aggressive or benign disease course, respectively. It is not known whether patients carrying mutations in these genes exhibit network-level abnormalities similar or different to those seen in sPD.
Method: 54 genotyped subjects (14 PD-LRRK2, 12 PD-GBA, 14 sPD, 14 HC) were closely matched for symptom duration and severity, as well as demographics. Using graph theory and metabolic brain imaging with [18F]-FDG-PET, we evaluated compensatory responses at the network level. Disease-related brain networks were studied with spatial covariance mapping [1]. Graph theory was employed to assess differences in nodal organization within the PD-related pattern (PDRP). We used a modularity maximization algorithm [2] to partition the PDRP space into a metabolically active core community (Module A) and an underactive peripheral community (Module B). We then used connectivity measurements in HC to assess gain and loss of connections within and between PDRP communities in each group.
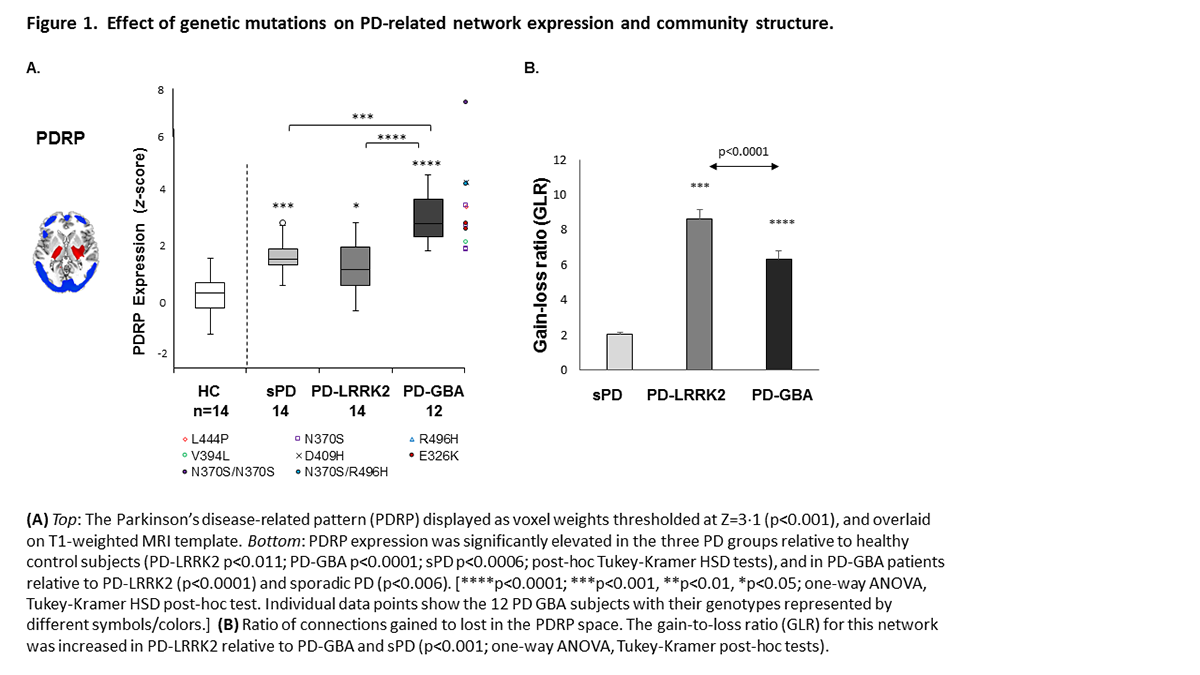
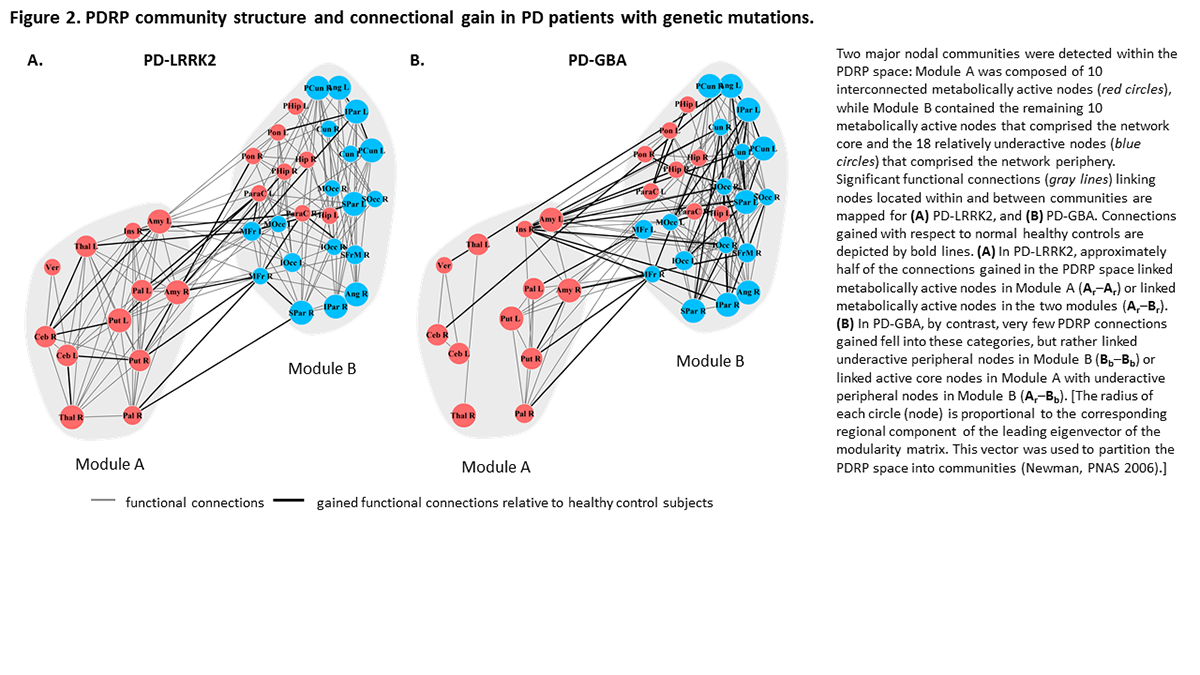
Results: PDRP expression levels were abnormally elevated in genotypic and sPD; we found no evidence for specific genotype-related networks (Fig.1A). The gain-to-loss ratio in the PDRP space, a measure of network-level functional compensation was significantly increased in PD-LRRK2 and PD-GBA relative to sPD (Fig.1B). In PD-LRRK2, a large portion of the gained connections linked active core nodes in Module A, such as those between the cerebellum, thalamus, and putamen. Functional compensation was lower in PD-GBA relative to PD-LRRK2, but higher than sPD, with gain in connections between the underactive peripheral nodes in Module B(Fig.2).
Conclusion: Genotype-specific alterations in functional connectivity were present in PD-LRRK2 and PD-GBA but not in sPD and HC. The changes in PD-LRRK2 reflect efficient network-level compensatory responses involving the PDRP core, particularly cerebello-thalamo-striatal pathways. The changes in the more aggressive PD-GBA genotype, involved increased cortico-cortical connectivity at the network periphery – a less efficient form of compensation.
References: 1. Spetsieris PPG, Eidelberg D. Scaled subprofile modeling of resting state imaging data in Parkinson’s disease: Methodological issues. Neuroimage. 2011;54(4):2899–914. 2. Newman MEJ. Modularity and community structure in networks. PNAS. 2006;103(23):8577–82.
To cite this abstract in AMA style:
K. Schindlbeck, A. Vo, N. Nguyen, C. Tang, M. Niethammer, V. Dhawan, V. Brandt, R. Saunders-Pullman, S. Bressman, D. Eidelberg. Genotype influences circuit compensation in Parkinson’s disease [abstract]. Mov Disord. 2019; 34 (suppl 2). https://www.mdsabstracts.org/abstract/genotype-influences-circuit-compensation-in-parkinsons-disease/. Accessed February 8, 2026.« Back to 2019 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/genotype-influences-circuit-compensation-in-parkinsons-disease/