Session Information
Date: Monday, September 23, 2019
Session Title: Huntington’s Disease
Session Time: 1:45pm-3:15pm
Location: Agora 3 West, Level 3
Objective: This preliminary study was designed to investigate the gene expression profile by RNA-seq in HD patients with depression and without depression, and between subjects with HD and healthy controls.
Background: Besides motor symptoms, Huntington’s disease (HD) is marked by cognitive impairment and behavioral changes. Among several psychiatric symptoms, depression is of great relevance as it is considered a significant burden for both individuals with HD and their caregivers [1]. In addition, the presence of depression is a predictor of suicidal ideations, attempts and completion [2]. Despite the high frequency and significance of depressive symptoms in HD, there is a lack of understanding of depression in HD.
Method: We performed RNA-seq analysis of peripheral blood of HD patients with depression (n = 8), HD patients with no depression or history of depression (n=8) and healthy control subjects (n =8), all matched for age and sex. Preprocessing of data and between-group comparisons were calculated using DESeq2. The Wald test was used to generate p-values and log2 fold changes. Genes with a p-value < 0.05 and absolute log2 fold change > 1 were called as differentially expressed genes. In addition, we clustered differentially expressed genes by their gene ontology (GO) using GeneSCF and the enrichment of GO terms was tested by Fisher exact test.
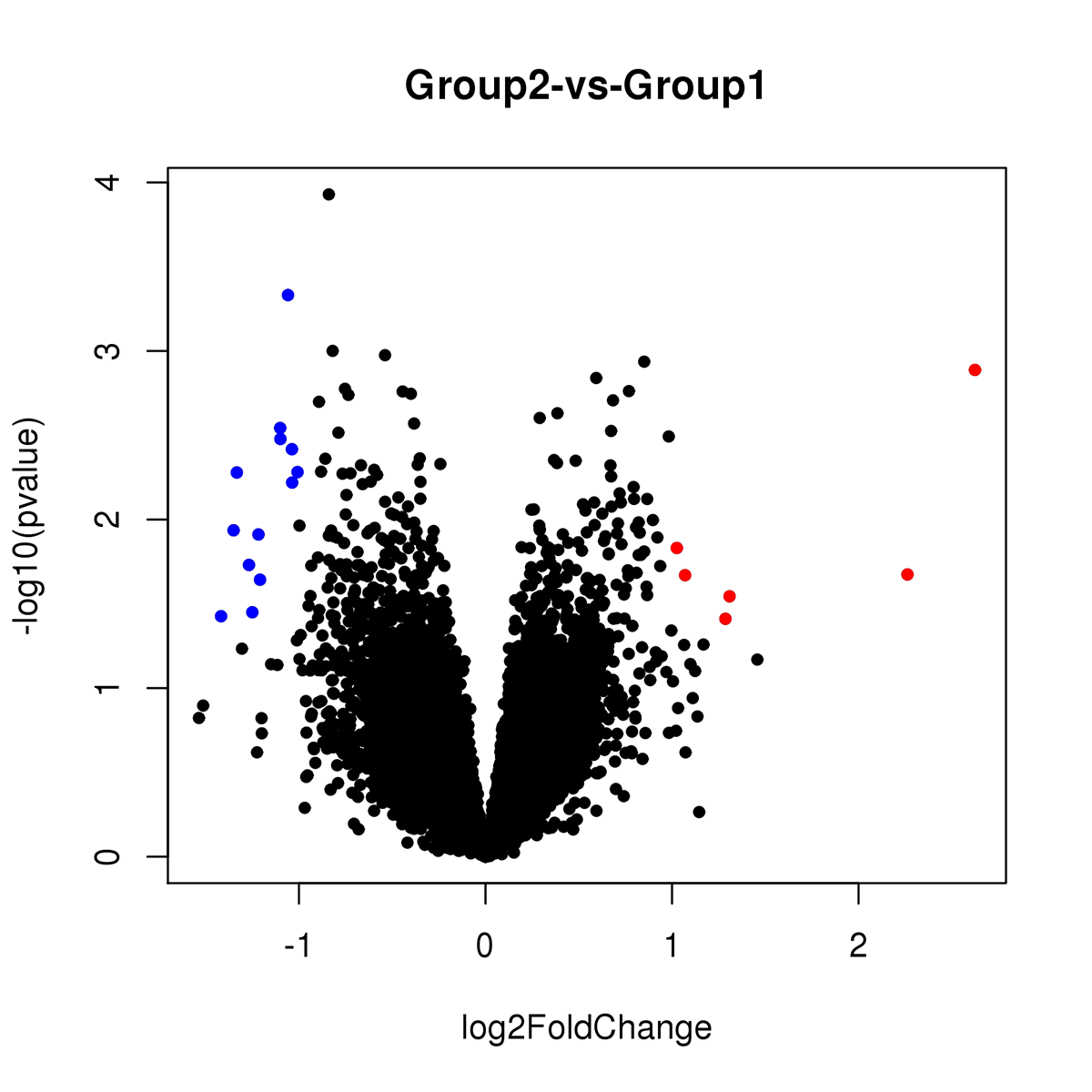
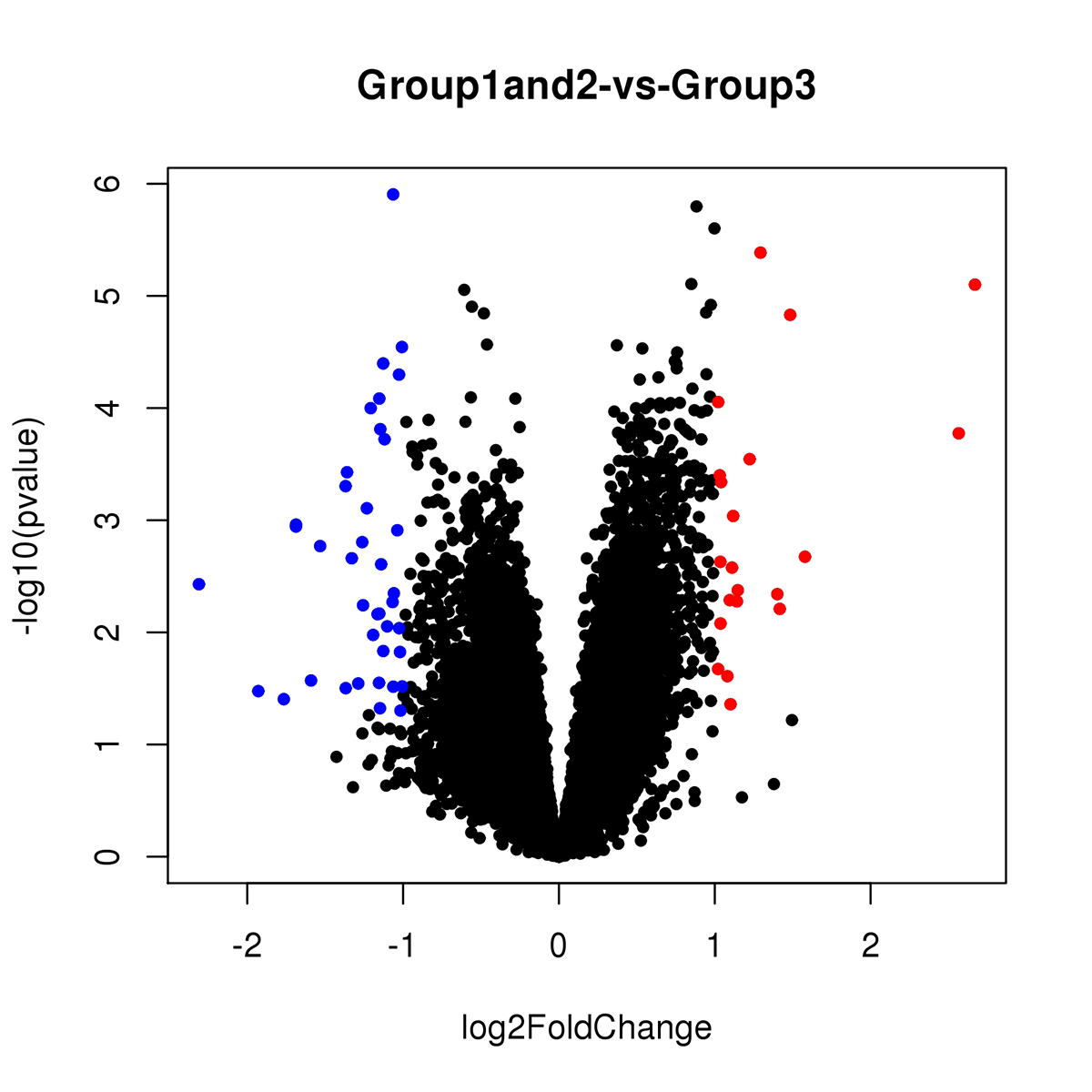
Results: In this exploratory analysis, we found 19 genes differently expressed between HD patients with depression and HD without depression. Among these genes, 6 were upregulated and 13 were downregulated (Figure 1). According to the GO analysis, these genes are involved in several pathways, such as apoptotic process, immune response, cell differentiation, establishment of mitochondrion localization, among others. In the analysis between all HD patients (n = 16) and healthy controls, we found 60 genes differently expressed, of which 21 were upregulated and 39 were downregulated (Figure 2). According to the GO analysis, these genes are involved in neuron migration, neuron cell-cell adhesion, brain development, nervous system development, among others.
Conclusion: Based on our preliminary results, we suggest that different genetic pathways are involved in depressive phenotype in patients with HD. These information may lead to a better understanding of depression in HD, a more accurate diagnosis, and more effective treatments.
References: [1] Ready RE, Mathews M, Leserman A, Paulsen JS. Patient and caregiver quality of life in Huntington’s disease. Movement disorders: official journal of the Movement Disorder Society. 2008;23(5):721-6. [2] Wetzel HH, Gehl CR, Dellefave-Castillo L, Schiffman JF, Shannon KM, Paulsen JS, et al. Suicidal ideation in Huntington disease: the role of comorbidity. Psychiatry research. 2011;188(3):372-6.
To cite this abstract in AMA style:
G. Colpo, N. Rocha, E. Furr Stimming, A. Lucio. Gene Expression Profiling of depression in Huntington’s disease [abstract]. Mov Disord. 2019; 34 (suppl 2). https://www.mdsabstracts.org/abstract/gene-expression-profiling-of-depression-in-huntingtons-disease/. Accessed February 1, 2026.« Back to 2019 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/gene-expression-profiling-of-depression-in-huntingtons-disease/