Session Information
Date: Tuesday, June 6, 2017
Session Title: Parkinson's Disease: Pathophysiology
Session Time: 1:45pm-3:15pm
Location: Exhibit Hall C
Objective:
To determine if metabolomic analysis – untargeted profiling of small-molecule (<1.5 kDalton) biochemical constituents – can yield useful diagnostic biomarkers of PD using CSF and plasma specimens.
Background:
As PD is associated with systemic mitochondrial and other metabolic alterations, metabolomic analysis has the potential to recognize PD-specific patterns capable of offering offer diagnostic correlations and insights into the disease process.
Methods: Biospecimens came from BioFIND study* participants, whose PD subjects met rigorous diagnostic criteria and were symptomatic for at least 5 years. Standardized biospecimen procedures were used for collecting CSF and plasma from 50 HCs matched to 50 PD subjects. Assays run in duplicate involved a platform of ultrahigh-performance liquid chromatography linked to with tandem mass spectrometry**. A spectral reference library provided chemical identifications. Data underwent intensive curation and quality-control measures. Each analyte was tested individually for association with PD using the t test. False-discovery rate (FDR)-adjusted p-values were calculated. Least Absolute Shrinkage and Selection Operator (LASSO) logistic regression was used to identify a multi-metabolite profile for discriminating between PD and HCs.
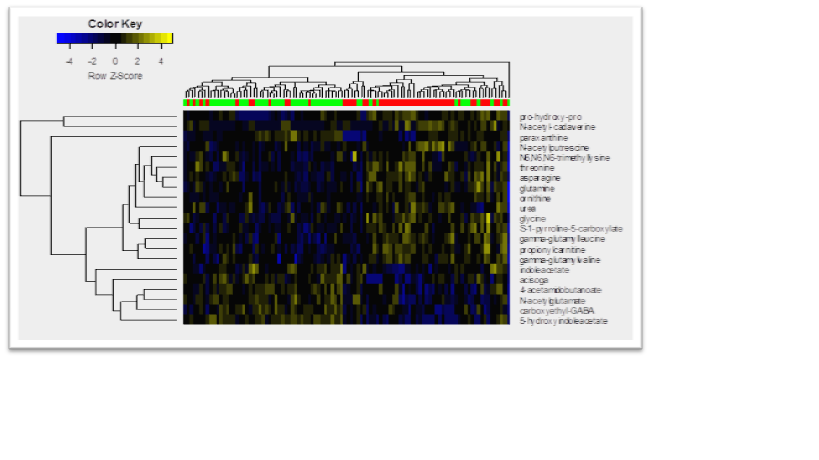
Results: The LASSO logistic regression selected 23 biochemicals to distinguish PD from HCs with area-under-the-receiver-operating-curve of 89.7%. With optimal cutoff, LASSO achieved 100% sensitivity and 96% specificity (with 96% positive and 100% negative predictive values).Ten-fold cross-validation gave 84% sensitivity and 82% specificity (with 82% positive and 84% negative predictive values).The top 3 markers were the putrescine precursor ornithine, the polyamine metabolite N-acetylputrescine, and an isobar indicating either N-acetylglucosamine or N-acetylgalactosamine. In the univariate analysis (Figure), 21 CSF compounds were differentially expressed between PD and HCs (FDR <0.05). Metabolic pathway analysis of the LASSO-selected biomarkers was not informative. In contrast to CSF, plasma metabolomic analysis did not distinguish PD from HCs.
Conclusions: Metabolomic profiling of CSF provided strong prediction of PD versus HCs. Biomarkers discovered by this analysis may be useful at enhancing diagnosis of PD and may provide better understanding of its etiology – for example, the top markers selected by LASSO are components of cellular pathways that have been implicated in aggregation of α-synuclein. The metabolomic analytical system used in this research has high through-put capabilities and standardization that is optimal for replication and inter-assay comparisons.
References: * Kang UJ, Goldman JG, Alcalay RN, et al. The BioFIND study: characteristics of a clinically typical Parkinson’s disease biomarker cohort. Mov Disord 2016;31:924-932. ** LeWitt PA, Li J, Lu M, Guo L, Auinger P, Parkinson Study Group – DATATOP Investigators. Metabolomic biomarkers as strong correlates of Parkinson disease progression. Neurology (in press, 2017)
To cite this abstract in AMA style:
P. LeWitt, J. Li, M. Lu, K.-H. Wu, Y. Zhou, I. Datta, L. Guo, B. Investigators. Metabolomic biomarkers strongly differentiate PD from healthy controls (HCs) in BioFIND study* specimens [abstract]. Mov Disord. 2017; 32 (suppl 2). https://www.mdsabstracts.org/abstract/metabolomic-biomarkers-strongly-differentiate-pd-from-healthy-controls-hcs-in-biofind-study-specimens/. Accessed July 18, 2025.« Back to 2017 International Congress
MDS Abstracts - https://www.mdsabstracts.org/abstract/metabolomic-biomarkers-strongly-differentiate-pd-from-healthy-controls-hcs-in-biofind-study-specimens/